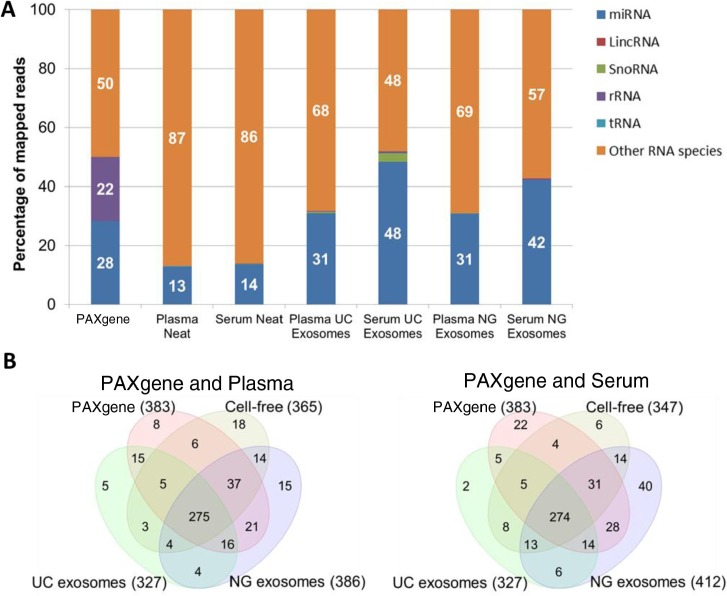
Fig. 4.
Percentage and number of known miRNA species detected in intracellular, cell-free and exosomal samples by NGS. Raw reads were aligned to the human genome (HG19) and mapped to miRBase V.20 and other small RNA from Ensembl Release 74 followed by normalization of raw reads to RPM. The mean of reads per miRNA (n=3) was calculated. A) Percentage of total reads mapped to non-coding small RNA and other RNA species identified by deep sequencing. B) Venn diagrams showing unique and common miRNA detected in different components of blood. miRNA with read counts >5 reads per million were shown for comparison. miRNA identified in this study was uploaded to http://www.microvesicles.org (17).