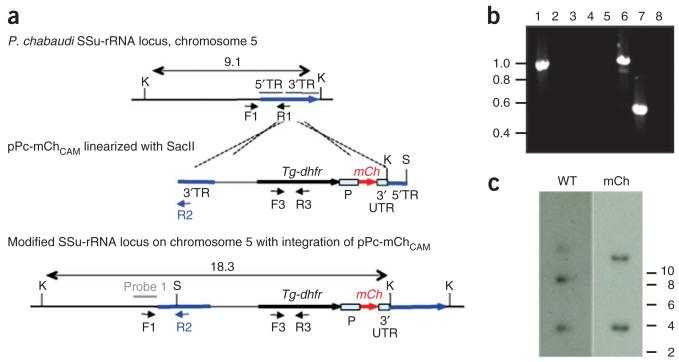
Figure 4. An example of vector design and genotypic analysis of transformed P.c. chabaudi parasites.
Parasites were transfected with plasmid pPc-mChCAM that is designed to achieve targeted integration into the small subunit ribosomal RNA (SSu-rRNA) loci on P.c. chabaudi chromosomes 5 (C5) or 6 (C6). (a) The wild-type (WT) integration locus, plasmid construct and resultant genetically modified locus after vector integration by single crossover recombination. The vector contains the gene encoding the drug-selectable marker, Tg-dhfr, the fluorescent marker mCherry (mCh) between the constitutive EF1α promoter (P), 3′ untranslated sequence (3′ UTR) and target sequences (TR) for integration. The construct was linearized with SacII (S) before transfection. (b) PCR analysis of genomic DNA showing correct integration of pPc-mChCAM into the SSu-rRNA locus on P.c. chabaudi C5. Primers C5-F1 (5′-GTAAATAAAAGCACTACTAATAAGTGTG-3′) and C6-F1 (5′-TGTAAATAAAAGCACTACTAATAAGTG-3′) are designed to anneal specifically to upstream regions of SSu-rRNA on P.c. chabaudi C5 or 6, respectively. Primer R1 (5′-TGGAGCCCTGTGATGATTC-3′) anneals with C5 and 6 SSu-rRNA sequences. Primer R2 (5′-AAGTAACGAGAAACCCAGTC-3′) anneals within the 38-bp region of pPc-mChCAM target sequence that differs significantly from the P.c. chabaudi WT SSu-rRNA sequence. Verification of the 5′ integration site in P.c. chabaudi C5; C5-F1/R2 amplify a product in mCherry-expressing P.c. chabaudi AS (mCherryPcc AS; lane 1) but not in wild-type P.c. chabaudi AS (Pcc AS; lane 2) DNA; C6-F1/R2 does not amplify a product in mCherryPcc AS (lane 3) or wild-type Pcc AS (lane 4) DNA. Parasites containing WT C5 SSu-rRNA sequences are not present; C5-F1/R1 amplify a product in mCherryPcc AS (lane 6) but not in wild-type Pcc AS (lane 5) DNA. Tg-dhfr is present in the genome of mCherryPcc AS (lane 7) but not in wild-type Pcc AS (lane 8) parasites (amplification with primers F3 (5′-ATGCATAAACCGGTGTGTCT-3′) and R3 (5′-CTACACGCGTGATGTACAGG-3′)). (c) Southern blot analysis of restricted genomic DNA to show integration of pPc-mChCAM. A probe designed against the upstream region of WT P.c. chabaudi C5 and C6 SSu-rRNA recognizes Kpn1 (K)-restriction bands of 9.1 and 4 kb, respectively. Integration of pPc-mChCAM into the C5 SSu-rRNA locus through single crossover recombination leads to shift in size from 9.1 to 18.3 kb.