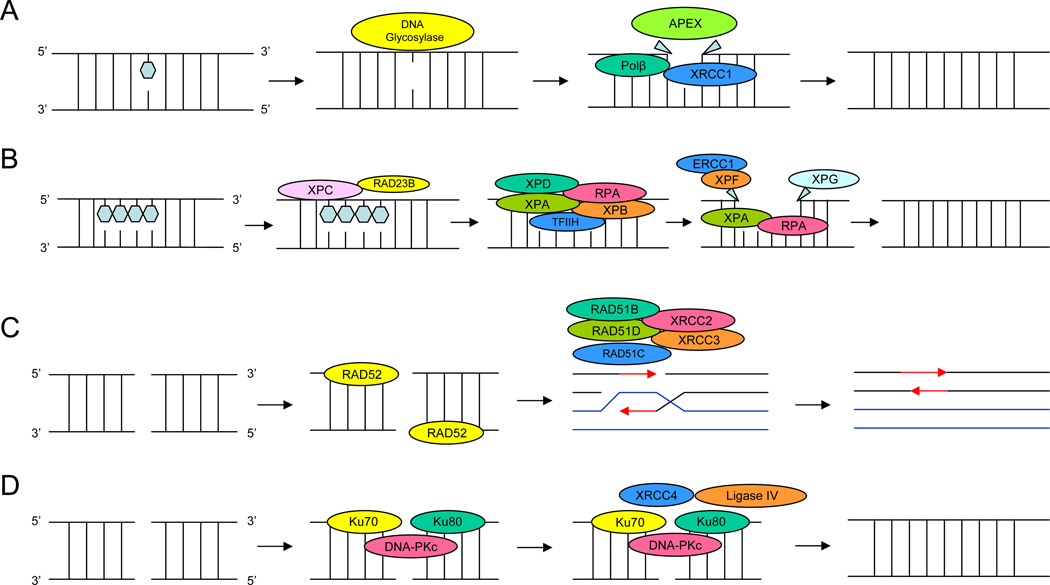
Figure 1.
1A: Base excision repair (BER) pathway targets DNA damaged during replication or by environmental agents. The single damaged base in DNA caused by endogenous metabolism or environmental oxidizing agents result in DNA adducts. Base excision repair involves removing the mutated base out of the DNA and repairing the base alone. 1B: Nucleotide excision repair (NER) is associated with the repair of bulky adducts induced by several suspected environmental prostate cancer carcinogens. The NER pathway is a complex biochemical process that requires at least four steps: (a) damage recognition by a complex of bound proteins including xeroderma pigmentosum complementation group C (XPC), XPA, and replication protein A (RPA); (b) unwinding of the DNA by the transcription factor IIH (TFIIH) complex that includes XPD(ERCC2); (c) removal of the damaged single-stranded fragment (usually about 27–30 bp) by molecules including an ERCC1 and XPF complex and XPG; and (d) synthesis by DNA polymerases. 1C: Double-strand breaks are produced by replication failure or by DNA damaging agents. Two repair pathways exist to repair double strand breaks. The homologous recombination repair relies on DNA sequence complementarity between the intact chromatid and the damaged chromatid as the bases of stand exchange and repair. 1D: The non-homologous end-joining repair pathway requires direct DNA joining of the two double-strand-break ends.