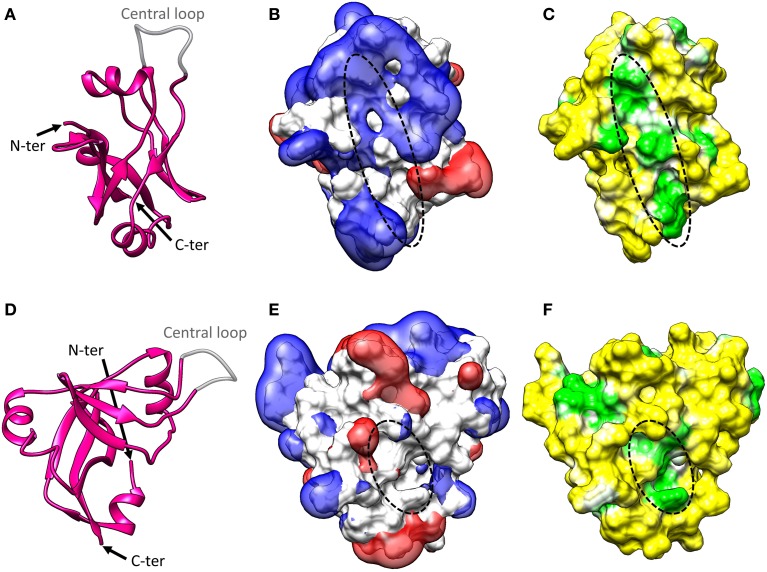
Figure 3.
SmpB structure. (A) Cartoon representation of the crystal structure of Thermus thermophilus SmpB (PDB entry: 1WJX). The N-terminal end, C-terminal end, and central loop are indicated. SmpB adopts an oligonucleotide-binding fold (OB fold) with a central β-barrel and three flanking α-helices. The C-terminal tail is unstructured in solution but folds into a fourth α-helix once SmpB is inserted into the ribosome. The central loop is disordered in the crystal, suggesting that it must be flexible when the protein is alone. (B) Electrostatic potential of SmpB. Two isocontours at −1 V (red) and +1 V (blue) are represented with the solvent-accessible surface of SmpB (white). The potential was calculated using the APBS program (Baker et al., 2001) with CHARMM force field parameters and an ionic strength of 50 mM. The primary tmRNA binding site interacting with the TLD (indicated with a dotted line) is surrounded by a strong electropositive field. (C) Molecular hydrophobicity potential projected on the solvent-accessible surface of SmpB. The potential was computed with the Platinum server using Ghose force field parameters (Pyrkov et al., 2009). The hydrophobicity scale is green-white-yellow, with yellow representing the most hydrophilic regions and green the most hydrophobic. The primary tmRNA binding site interacting with the TLD (indicated with a dotted line) is formed by a deep hydrophobic patch. (D–F) Side views of the information presented in (A–C). Note that in (E,F), a dotted line indicates the secondary tmRNA binding site that interacts with the nucleotides upstream from the resume codon after translocation of tmRNA-SmpB into the ribosomal P-site.