Abstract
Early diagnosis of fungal infection is critical to effective treatment. There are many impediments to diagnosis such as a diminishing number of clinical mycologists, cost, time to result, and requirements for sensitivity and specificity. In addition, fungal diagnostics must meet the contrasting needs presented by the increasing diversity of fungi found in association with the use of immunosuppressive agents in countries with high levels of medical care and the need for diagnostics in resource-limited countries where large numbers of opportunistic infections occur in patients with AIDS. Traditional approaches to diagnosis include direct microscopic examination of clinical samples, histopathology, culture, and serology. Emerging technologies include molecular diagnostics and antigen detection in clinical samples. Innovative new technologies that use molecular and immunoassay platforms have the potential to meet the needs of both resource-rich and resource-limited clinical environments.
New generations of fungal diagnostics must reconcile the needs of patients in developed countries with those in resource-limited settings. Emerging technologies that use molecular and immunoassay platforms may meet these needs.
There has been an enormous increase in the frequency and severity of fungal infection in recent years. This increase has been driven in a large part by two factors. First, the global AIDS epidemic has fostered the emergence of life-threatening infections by the opportunistic fungi Cryptococcus neoformans and Pneumocystis jiroveci and by regional endemic fungi such as Histoplasma capsulatum and Penicillium marneffei. These infections occur most often in resource-limited countries in Africa, South America, and Southeast Asia. Second, advances in medical care and treatment have led to increases in the number of opportunistic infections in patients who are immunocompromised by way of treatment with immunosuppressive drugs or chemotherapy, or who are infected in the course of extended critical care. These infections occur most often in the setting of advanced medical care. The result is patients with infections that are difficult to diagnose and difficult to treat.
Diagnosis of invasive fungal disease (IFD) is challenging because current diagnostic methods lack sensitivity and specificity, or take too long to yield a result to be clinically useful. Such limitations have consequences; delayed diagnosis leads to delayed treatment. Speed to diagnosis is a key risk factor in patient outcomes (Barnes 2008). Diagnosis of fungal infection is further complicated by problematic developments in the field of medical mycology. First and foremost is the loss of senior mycology experts in the field who were trained in classical mycology, which has created crises-level problems in clinical mycology (Steinbach et al. 2003). This problem has been compounded over the last 30 years as the spectrum of fungi-causing infections has exploded owing to AIDS and the use of highly immunosuppressive agents for treatment of a variety of diseases. These patients are susceptible to infections from fungi rarely seen, or never reported as a human pathogen, which can cause identification problems for even the most experienced mycologists. Whereas mycologists in the past needed to be able to identify ∼50 commonly encountered fungi, and ∼300 total fungi that were pathogenic for humans, the number of potential fungal pathogens is likely many times what is described in textbooks, and will continue to grow as the severely immunosuppressed patient population continues to grow (Ajello and Hay 1998; Collier et al. 1998; Andrew et al. 2009).
Diagnosis of fungal infection has relied primarily on methods such as direct microscopic examination of clinical samples, histopathology, and culture. Such approaches are dependent on personnel with relatively high levels of specific mycology training. The growth in the number of fungi that clinical mycologists must identify has forced investigators to develop and apply new methods for fungal identification that go beyond classical phenotypic methods. As a consequence, there is an increased emphasis on the use of molecular methods and antigen detection as surrogates for culture in diagnosis of fungal diseases.
CULTURE, DIRECT MICROSCOPY, AND HISTOPATHOLOGY
Culture, direct microscopy, and histopathology have been the foundation for diagnosis of fungal infection for many decades. Microscopy, histopathology, and use of fungal-specific stains play important roles in diagnosis of infection by C. neoformans, P. jirovecci, Candida spp., Aspergillus spp., H. capsulatum, Blastomyces dermatitidis, Coccidioides immitis, Sporothrix schenckii, Paracoccidioides brasiliensis, and the Mucorales. Sensitivity of microscopy for diagnosis of fungal infection varies with the individual agent, the source and quality of the specimen, and the skills and experience of the laboratorian. Finally, diagnosis of invasive fungal infection by direct microscopy and histopathology may require the use of biopsies of deep tissues, which poses a risk to those patients who are most susceptible to invasive disease.
Culture from a clinical sample is the gold standard for diagnosis of fungal infection. Culture has the advantage of yielding the specific etiological agent if positive. Moreover, culture allows for susceptibility testing. However, use of culture for diagnosis of IFD has significant limitations. Culture may take many days to a result with several of the filamentous fungi. In the case of disseminated candidiasis, blood culture (1) may miss ≥50% of patients with documented disease (Fraser et al. 1992; Ostrosky-Zeichner and Pappas 2006; Ostrosky-Zeichner 2012), (2) may only become positive late in infection (Ellepola and Morrison 2005), and (3) typically takes 24–72 h for identification of Candida in a clinical sample—too long for early treatment. Positive blood culture is rare in invasive aspergillosis and is most often owing to environmental contamination (Kontoyiannis et al. 2000). Recovery of H. capsulatum from sputum of patients with acute pulmonary histoplasmosis ranges from 10% to 15%; however, in cavitary histoplasmosis, sputum cultures are positive in up to 60% of patients (Deepe 2010). In patients with pulmonary blastomycosis, sputum culture or culture of specimens obtained by bronchoscopy has a high yield (86% per patient for sputum culture and 92% for broncoscopy) (Chapman and Sullivan 2010). Culture of Coccidioides spp. is complicated by the biosafety hazard associated with culture of the mycelial form. Finally, identification of less common fungi that may cause opportunistic infections requires a high level of expertise on the part of laboratory personnel.
SEROLOGY
Serologic tests for patient antibodies have been useful for non-culture-based diagnosis of fungal infection since the middle of the last century. Serology is of greatest value in diagnosis of endemic mycoses. Available technologies include immunodiffusion (ID), complement fixation (CF), and enzyme immunoassay (EIA).
CF and immunodiffusion are the most common serologic tests for diagnosis of histoplasmosis. The ID test detects precipitating antibodies to Histoplasma H and M antigens. Serologic testing for histoplasmosis is most useful if an increase in CF titer is observed between acute and convalescent sera in acute histoplasmosis (Deepe 2010). High titers may be observed with chronic pulmonary or disseminated histoplasmosis (Lindsley et al. 2006).
Serological testing plays an important role in diagnosis of coccidioidomycosis, particularly in patients who may not be able to produce a sputum sample, for example, primary infection, or when samples are often negative, for example, coccidioidal meningitis (Galgiani 2010). Depending on the antigen used in the test, the qualitative ID test will determine the presence of coccidioidal IgM with a result that is similar to a tube precipitin test (IDTP) or coccidioidal IgG that detects antibody recognized by the CF test (IDCF). Detection of IgM is useful in diagnosis of acute primary coccidioidomycosis in which the sensitivity may be >80% (Saubolle et al. 2007). CF detects IgG antibodies. IgG antibodies are produced during the convalescent phase of disease or during chronic infection. CF is more sensitive than IDCF and provides quantitative results. A commercially available EIA can be used to detect IgM or IgG antibodies.
There are many advantages to the use of serology for diagnosis of invasive fungal infection. First, results may be positive when culture results are negative or samples are difficult to obtain. Second, if positive, serological results may reduce the need for culture of potentially hazardous fungi, for example, Coccidioides spp. Finally, serology is a minimally invasive sample, which lowers barriers to testing. Disadvantages of serology include sometimes low levels of sensitivity and specificity. A negative serologic test should not exclude the presence of fungal infection. Some tests, particularly CF, are time consuming and require trained personnel. Immunocompromised patients may show a reduced antibody response, which would dramatically reduce the value of serologic assays (Tobón et al. 2005). Interpretation of serological results may be confounded by the inability of serology that measures IgG to distinguish between current or previous infection. False positives may occur with some tests in the setting of other endemic fungal infections. Finally, sensitivity is dependent on the type of disease and the timing of testing relative to the disease process, for example, early versus late.
MOLECULAR DIAGNOSTICS
In the field of clinical mycology, no area is advancing faster than the application of modern molecular tools for the identification of fungi. The advancement of this area has been driven in large part by the rapid accumulation of protein and DNA sequence data, which continues unabated and accelerates with each new advance in technology, and the growing need to identify a broader range of fungi. Importantly, molecular methods for fungal diagnosis and identification directly address the declining numbers of clinical mycologists because they are not dependent on classical phenotyping methods. Similarly, molecular methods have the power to identify the increasing numbers of fungi found to produce disease in humans and animals.
Non-Culture-Based Molecular Diagnostic Methods
Molecular identification methods form a subset of diagnostic methods that do not necessarily need live fungal cells for success. Molecular methods for fungal identification generally work best when pure cultures are available. However, because polymerase chain reaction (PCR) plays a role in many molecular identification methods, molecular identification can work in the absence of live cells if template nucleic acid is available in patient specimens, including fixed tissue.
PCR is a central component for many molecular methods, either as the main diagnostic strategy or as one of the preliminary steps in the diagnostic assay. Consequently, diagnostic PCR encompasses a number of different approaches. The simplest consists of conventional PCR in which species-specific primers that have been designed based on existing sequence or data, are used to amplify fungal DNA from clinical specimens. The readout generally consists of the presence or absence of a band, with the size of the band often being a secondary factor in identification. This type of PCR is not FDA approved and although simple and inexpensive, can be subject to wide intra- and interlaboratory variation. There also must be some prior suspicion about the identity of the isolate, which the PCR reaction will confirm. Nonetheless, it is still used in research laboratories that occasionally may need to perform diagnostic studies on select organisms and can be suitable for minimally equipped laboratories if there is a stable power supply to run the thermocycler.
Further discriminatory power can be added to conventional PCR techniques by using restriction enzymes to digest the PCR products, which are resolved on a gel and visualized as the final step in the assay. The presence or absence of a fragment and their various sizes can then be used to make an identification. This method was used extensively in early fungal taxonomy (de Hoog et al. 2000). Unfortunately, owing to the great variability of PCR and the confirmatory nature of the technology (results are positive or negative, depending on the suspected organism), it is not widely used in the clinical laboratory. Importantly, conventional PCR serves as a confirmatory assay in which a positive reaction simply confirms the identity of the suspected fungus. The confirmatory nature of the assay is derived from the need to design primers that anneal to specific target sequences previously identified from a known organism. However, degenerate primers can expand this identification to genus or higher taxonomic levels, and if the product is to be sequenced, special strategies are used in primer design and target selection to greatly expand the number of fungi that can be amplified.
In contrast to conventional PCR, which requires gel electrophoresis of ethidium bromide stained PCR products as the final readout, real-time PCR uses fluorescent dyes to enhance specificity through either a nonspecific DNA-binding dye, that is, SYBR green, or a specific fluorescently labeled probe directed to a target sequence lying within the amplicon. There are various chemistries for probe labeling and fluorescent detection (i.e., Taqman, Molecular Beacons, etc.), as well as numerous thermocycler platforms (i.e., LightCycler, GeneAmp, etc.), which, depending on the combination, can have various levels of throughput. The application of fluorescent probes to PCR has made this technology suitable for the clinical laboratory and even has nanolevel applications that enable the identification of multiple organisms using different matrices.
A number of assays have been FDA approved for nonfungal identification (i.e., influenza, methicillin-resistant Staphylococcus aureus), whereas only a few have been approved for fungal identification, such as the FilmArray Blood Culture Identification (BioFire Diagnostics, Inc.), which is a PCR-based reaction performed on positive blood cultures and detects mainly Candida spp. Because PCR-based assays include an amplification step of a specific nucleic acid target, they can be performed on specimens that are potentially contaminated with human tissue or fluids, which is a tremendous advantage of PCR because a time-consuming outgrowth period is not needed and the assay can often be completed in a few hours. Furthermore, PCR can be performed on fixed tissue with some success, although these templates come with special challenges (Dannaoui et al. 2010). Like conventional PCR, real-time PCR generally is confirmatory in that a suspicion of the organism identity is established before performing the PCR reaction. Additionally, although multiplexable, a problem with probe-based diagnostic assays is that each assay must contain all probes for the organisms it can detect, which can entail substantial reagent costs, even when performed on a nanoscale.
Culture-Based Molecular Diagnostic Methods
Establishing a pure culture of a suspected microbial sample isolated from a clinical specimen has always been the gold standard of diagnostic microbiology, regardless of the downstream diagnostic assay that is applied for the ultimate identity of the organism. Although in many instances it is not possible to obtain a pure, viable culture, virtually all diagnostic assays work best if the assay initiates from a pure culture. Pure cultures ensure there is enough material to perform the assay, enable repeat assays in the case of failure, allow for sending the isolate to alternate sites where the assay can be performed, and/or allow additional unrelated assays to be performed if confirmation or a more discriminatory strategy is needed. Culture-based methods historically have been phenotype driven, in which yeasts were identified biochemically and molds were identified based on morphological features. These methods are still important, frontline diagnostic methods. However, with the application of molecular biology to fungal taxonomy and phylogeny, it has become clear that molecular biology needs to be part of diagnostic mycology. As a result, the major molecular taxonomic tool, ribosomal sequencing, is now the major molecular tool for fungal identification.
Sequencing of fungal ribosomal targets is an attractive diagnostic method for a number of reasons. First, fungal ribosomes and most eukaryotic ribosomal genes are multicopy in nature, which increases detection sensitivity during PCR amplification because there are more target sequences. Second, the organization of these loci in fungi places multiple conserved ribosomal (18s, 5.8s, and 28s) subunit genes in close proximity, which offers conserved PCR primer sites that are positioned such that multiple target sites are close enough to yield PCR products. In fact, the conserved nature of the subunits, and their primer annealing sites, makes PCR and sequence identification possible, enabling virtually any unknown fungus to be amplified with universal primers targeted to these regions (White et al. 1990; Kurtzman and Robnett 1997). Third, the overall organization of this region confers variability owing to the fact that variable regions separate the key ribosomal units. These regions, called the internal transcribed spacer (ITS) sequences, consist of two regions (ITS1 and ITS2) that are not part of the fungal ribosome and are spliced out after transcription (Fig. 1). Their presence confers the sequence variability that makes rDNA sequencing the most powerful nucleic acid–based diagnostic method available. In addition to the variable ITS1 and ITS2 regions, a third variable region exists within the large 28s ribosomal subunit called the D1/D2 region. All three regions are informative owing to their variable nature, which can yield genus-specific and species-specific identifications. Finally, sequence data generated from an unknown fungus can be used to search the public databases, such as GenBank, using the web-based BLASTn algorithm.
Figure 1.
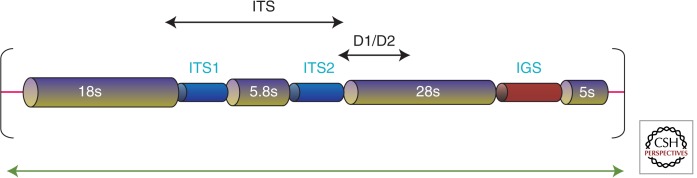
Ribosomal subunit organization in eukaryotes with variable regions. Eukaryotic ribosomal subunit genes are typically organized in repeats of the 18s rDNA (small subunit), ITS1 (internal transcribed spacer region 1), 5.8s rDNA, ITS2 (internal transcribed spacer region 2), and the 28s rDNA (large subunit). Conserved priming sites exist at the end of the 28s subunit and the beginning of the 28s subunit, and at the end of the D1/D2 region within the 28s subunit. Additional sites exist within the 5.8s subunit and throughout the large and small ribosomal subunits. The variable regions, which can provide information that can discriminate to the species level, depending on genus, include ITS1, ITS2, and D1/D2. The ITS region can be covered in a single PCR reaction and by double-stranded sequencing, and is roughly 450–750 bp in length, depending on species. The D1/D2 region is in a similar size range.
Database searches must be performed with extreme caution owing to the public nature of the database and high frequency of erroneous deposits (Bidartondo 2008). Nevertheless, there are hundreds of thousands of fungal sequences deposited within GenBank that can serve as potential identification data. It is the wide array of different fungal species that makes GenBank so powerful. It is unlikely that any fungal identification platform will be as broad in terms of the number of potential species that can be identified using DNA sequences because of the public nature of GenBank, length of time sequences have been deposited into the database, and widespread use of sequencing as a basic molecular technique. In fact, most journals that publish papers that include sequence information require a GenBank deposit of the data. Importantly, global access to GenBank ensures that fungal sequences from all over the world are regularly deposited, which serves to increase the diversity of information in the database. Unfortunately, the unedited nature of GenBank has always been problematic for fungal identification, which has resulted in the development of both commercial and publicly curated, closed databases.
Although most of the taxonomically significant sequences are ribosomal in nature, for some fungi, there is not enough specificity in the ribosomal loci to discriminate different species. In this case, alternate sites based on conserved structural genes have been developed and in many cases are standard depending on the presumed identity (Table 1) (Petti et al. 2008). However, to generate these sequences, specific primers must be used, which requires some information about the unknown fungus to guide primer selection. Some genes such as tubulin have primers that amplify across families (Glass and Donaldson 1995).
Table 1.
Alternate non-rDNA loci used for molecular identification
| Locus | Genera/order | References |
|---|---|---|
| β-Tubulin | Aspergillus, Penicillium, Phoma, Pseudallescheria, Chaetomium, Phaeoacremonium, Sporothrix, Paecilomyces | Glass and Donaldson 1995 |
| Preribosomal-processing protein | Aspergillus, Penicillium | Houbraken and Samson 2011 |
| Minichromosome maintenance protein | Aspergillus | Sugui et al. 2012 |
| Translation elongation factor 1α | Fusarium, Mucorales, Beauveria, Cordyceps, Trichoderma, Alternaria, Cladosporium, Phomopsis | O’Donnell et al. 1998, 2001; Abe et al. 2010 |
| Cytochrome oxidase 1 | Penicillium | Lasker 2006 |
| Calmodulin | Pseudallescheria, Sporothrix, Aspergillus | O’Donnell et al. 2000; Hong et al. 2006; Marimon et al. 2006 |
| Glyceraldehyde-3-phosphate dehydrogenase | Cochliobolus, Curvularia, Bipolaris | Berbee et al. 1999 |
| Endopolygalacturonase | Alternaria | Andrew et al. 2009 |
| RNA polymerase II | Schizophyllum, Alternaria, Penicillium, Aspergillus | Liu et al. 1999; Peterson 2008; Samson et al. 2011; Yilmaz et al. 2012 |
| Actin | Rhizopus, Stachybotrys, Trichophyton, Phaeoacremonium | Carbone and Kohn 1999; Gao and Takashima 2004 |
| Intergenic sequence | Trichosporon | Sugita et al. 2002 |
A requirement for sequence-based diagnostics is the need for bioinformatics skills to manipulate sequences and ultimately to search databases and interpret the results. This requirement has led to the development of alternate DNA-based identification technologies. One of these technologies (Luminex xMAP) uses color-coded microspheres that are specific for each analyte in a sample. In the case of fungal identification, this platform uses PCR of an unknown sample that can then be detected in a hybridization assay by binding to its corresponding bead in the assay. Detection and identification using this methodology is similar to the technology used in flow cytometry. Each assay is capable of detecting up to 100 different species in a single multiplex reaction (Preuner and Lion 2013). The advantage of this platform is that there are no downstream manipulations of data after the assay is complete. The multiplex capability also greatly exceeds a single PCR reaction; however, the true identity of the organism must be one of the 100 components of the multiplex.
Proteomics Profiling/Fingerprinting
The most popular and fastest growing non-nucleic acid sequence based molecular diagnostic assay for fungi is MALDI-TOF (matrix-assisted laser desorption/ionization time-of-flight). The technique generates species-specific spectra that provide a unique signature characteristic of the species. MALDI-TOF instrumentation consists of an ion source that transfers sample molecules into a gas phase, a mass analyzer that resolves ions based on mass-to-charge ratio, and a component that detects the ions (Croxatto et al. 2012). Samples are mixed with a matrix of small acidic molecules that crystallizes the specimen and facilitates ionization because the matrix absorbs energy in the range of the laser used for sample excitation. The TOF component consists of a tube that the excited ions travel through, with the transit time (time-of-flight) of individual ions providing the method for identification. The generated spectra are screened against a library of reference spectra, which correspond to individual species. The technology has already been commercialized for microbial identification, with instruments being available from Bruker Daltronics (MALDI Biotyper), Shimazu (AXIMA@SARAMIS), Andromas (Andromas), and bioMérieux (Vitek MS) (Bader 2013).
The strength of MALDI-TOF technology lies in the rapid sample analysis (minutes) and the absence of any downstream data manipulation. For many fungi, particularly many yeasts, sample prep is minimal. For other fungi, a rapid extraction with a solvent is all that is needed. These factors—the lack of downstream steps and simple sample preparation—combined with the accuracy and speed of this system make MALDI-TOF one of the most intriguing diagnostic options for fungal identification. Importantly, just as for sequencing, no prior suspicion of the true identity of the isolate is needed. If there is a reference spectrum in the library, there is a high probability that a correct identification can be made (Posteraro et al. 2013). Weaknesses of this system include the need for an existing spectral library to compare generated spectra to, and potential variability in results of unknown fungi if they are not grown under conditions similar to reference spectra in the library. Libraries are proprietary and instrument specific, although most are modifiable by users. There are capitalization costs for the instrumentation, and user skills need to be somewhat advanced. Portability is also not an option with this instrumentation.
Challenges to Molecular-Based Diagnostic Mycology
There remain a number of challenges preventing full implementation of a molecular-based platform as the main identification method in clinical microbiology laboratories. On the mycology side, the spectrum of fungi that can cause disease continues to grow and likely will be almost infinite as patient populations, depending on illness or treatment, can be so profoundly immunosuppressed that species never before seen as pathogenic for humans regularly appear in the clinical microbiology laboratory. Identifying these fungi using classical phenotypic methods is extremely difficult as laboratory staff generally do not have the necessary training or experience to arrive at an identification based on morphology for rare fungi. Biochemical identification is generally limited to commercial platforms that have panels of standard compounds, which are directed toward identifying common fungi found in a coded database depending on metabolism pattern of the panel. Furthermore, mycology is unique in that there is a special nomenclature system that takes into consideration whether or not a given isolate has a sexual state or not. Unfortunately, only the most experienced mycologists can navigate this system, which has become so complex that efforts are under way to reform it so that a simpler more stable naming system can be used (Hibbett and Taylor 2013). Beginning in 2013, consistent with the Amsterdam Declaration on Fungal Nomenclature, a unified effort will be made to insure that the one-fungus–one-name rule is implemented so that fungal nomenclature can be simplified (Hawksworth et al. 2011).
On the technical side, one of the simplest, yet longest running challenges impeding molecular diagnostics is a universal method for preparing sample templates. For sequence-based assays, fungi vary depending on their morphology in their resistance to cell lysis and ease of release of nucleic acid. There are countless methods that have been successful, ranging from enzymatic to chemical to physical; however, a “one-size-fits-all” method that could be commercialized has proven elusive. This problem also hinders development of strategies that could meet FDA approval because lack of consistency and varying requirements for technical input are problematic for certified laboratories. Additionally, the sheer number of fungi that a microbiology laboratory must be able to identify can preclude the use of assays, such as PCR, that can only identify one or a few fungi in the given assay. More open-ended assays, which theoretically can identify any species within a given platform’s reference libraries or databases, are more realistic because they could potentially replace all culture-based assays in the clinical laboratory, and can even be used across kingdoms. However, these assays tend to require expensive, stationary equipment that requires specialized skills to operate.
ANTIGEN DETECTION
Fungal polysaccharides or proteins may be shed into body fluids during the course of infection. If an antibody can be raised against such a shed antigen, an immunoassay can be constructed for antigen detection.
Cryptococcosis
Diagnosis of cryptococcal meningitis was the first application of antigen detection for diagnosis of fungal infection that received widespread clinical use (Bloomfield et al. 1963). Antibodies were raised in rabbits against whole cryptococcal cells and passively coated onto latex beads. Termed latex agglutination, the assay detected glucuronoxylomannan (GXM), the major capsular polysaccharide of C. neoformans. GXM is shed in large amounts into blood and cerebrospinal fluid (CSF) during the course of cryptococcal meningitis.
GXM occurs in four major serotypes: A, B, C, and D and a hybrid serotype AD. Before the use of molecular methods for classification of cryptococcal species, serotypes A, D, and A/D were C. neoformans; serotypes B and C were C. gattii (Bennett et al. 1977; Kwon-Chung and Bennett 1984b). With further study, C. neoformans var. grubii was identified to correspond to serotype A. C. neoformans var. neoformans corresponds to serotype D.
There is considerable variability in the geographic distribution of the cryptococcal species and their corresponding serotypes. Serotype A has a global distribution, and initial studies of cryptococcosis in patients with AIDS found a predominance of serotype A isolates, suggesting that serotype A was selectively infecting AIDS patients. However, there are several recent reports of serotype C in AIDS patients in sub-Saharan Africa where the frequency of serotype C has been reported to be as high as 14% (Karstaedt et al. 2002; Litvintseva et al. 2005; Thakur et al. 2009). C. neoformans var. neoformans (serotype D) has a global distribution, but clinical cases are concentrated in Europe (Kwon-Chung and Bennett 1984a; Dromer et al. 1996). Finally, C. gattii has gained recent prominence as the cause of an ongoing outbreak that began in Vancouver Island, British Columbia. Although there has not been an extensive serological characterization of isolates from the outbreak, molecular typing has found that almost all isolates are genetic variants of the VGII class on the basis of PCR fingerprinting; isolates of VGII produce capsules of serotype B (Litvintseva et al. 2011). Geographic variability in occurrence of cryptococcosis of different serotypes has consequences for diagnostic testing that targets cryptococcal antigen; assays for antigen must be able to detect GXM of all major serotypes.
A significant advance in testing for cryptococcal antigen was the development of an assay in lateral flow immunoassay (dipstick) format. Termed CrAg LFA, the assay is constructed from a cocktail of monoclonal antibodies that were formulated to be reactive with all GXM serotypes (Gates-Hollingsworth and Kozel 2013). The CrAg LFA is particularly well suited for use in resource-limited settings. The LFA requires no power or clean water, is inexpensive, requires no refrigeration, and can be performed by personnel with limited training. The LFA works well with serum or a drop of blood and provides a result in ∼10 min. As a consequence, patients can be treated at the time of an initial visit to clinic. The ease of use of the CrAg LFA also makes the test valuable for use in settings with advanced medical care.
Recent studies have found that CrAg testing can be used for prospective testing in asymptomatic patients at high risk for cryptococcosis. CrAg is present in serum for weeks to months before the onset of symptoms of cryptococcosis (Jarvis et al. 2009). As a consequence, a targeted screening program could identify subclinical infection in patients at greatest risk for cryptococcal meningitis. Specifically, HIV-infected patients can be screened for CrAg at the time of initial diagnosis of HIV/AIDS before starting antiretroviral therapy (World Health Organization Cryptococcal Working Group 2011). If a patient tests positive in this prospective screen, the patient can be preemptively treated to prevent infection from progressing to meningitis.
Galactomannan
In 1978, Lehmann and Reiss identified an antigen in serum from immunosuppressed rabbits infected with Aspergillus fumigatus (Lehmann and Reiss 1978). The investigators later found that the same antigen is released from growing hyphae (Reiss and Lehmann 1979). This antigen is galactomannan (GM), a polysaccharide present in the cell wall of most Aspergillus spp. A test for serum GM is now in widespread use for diagnosis of invasive aspergillosis (Platelia Aspergillus, Bio-Rad Laboratories). The test is an enzyme immunoassay that uses a rat monoclonal antibody that recognizes β (1→5)-linked galactofuranose (Stynen et al. 1992, 1995). Multiple studies have found the GM enzyme immunoassay (EIA) to be useful in diagnosis of invasive aspergillosis in neutropenic patients with cancer and recipients of stem cell transplants (Machetti et al. 1998; Maertens et al. 1999, 2001; Sulahian et al. 2001).
There are limitations to the GM immunoassay for diagnosis of invasive aspergillosis. First, there is considerable variability in reports of sensitivity and specificity (Machetti et al. 1998; Maertens et al. 1999, 2001; Pinel et al. 2003). Second, false positive reactions may occur owing to a variety of factors, including administration of β lactam antibiotics (Pinel et al. 2003) or infusion of gluconate-containing Plasma-Lyte (Petraitiene et al. 2011). Third, there is considerable cross-reactivity with other fungi producing disseminated infection (Huang et al. 2007; Wheat et al. 2007; Xavier et al. 2009).
Detection of H. capsulatum polysaccharide antigen in body fluids, especially urine, has been useful in presumptive diagnosis of histoplasmosis in patients with disseminated disease (Wheat et al. 1986). The antigen is a galactomannan (Connolly et al. 2007). Antigen can be detected in urine of ∼90% of patients with disseminated infection (Williams et al. 1994; Durkin et al. 1997). The sensitivity of antigen detection is greater in urine than in serum (Wheat et al. 2002). This is an antigen-capture ELISA in which polyclonal rabbit antibodies are used in both the solid-phase capture and fluid-phase indicator modes. Galactomannan recognized by immunoassay of urine in histoplasmosis patients is cross-reactive with polysaccharide antigen produced by several endemic mycoses (B. dermatitidis, P. brasiliensis, H. capsulatum var. duboisii, and P. marneffei) (Azuma et al. 1974; Wheat et al. 1997).
Immunoassays have been recently developed for detection of galactomannan from B. dermatitidis and C. immitis in urine and other body fluids. For B. dermatitidis antigen detection, antigenuria was detected in 90% of patients with culture- or histopathology-proven blastomycosis (Connolly et al. 2012). Specificity was 99% in healthy subjects and patients with nonfungal disease, but cross-reactions occurred in 96% of patients with histoplasmosis. For C. immitis antigen detection, antigenuria was detected in 71% of patients with more severe forms of coccidioidomycosis (Durkin et al. 2008). Additional studies are needed to assess the usefulness of galactomannan testing in other clinical forms of coccidioidomycosis.
Pan-Fungal Detection of β-Glucan
(1→3)-β-d-glucan (BG) is a polysaccharide component of most fungal cell walls. The polysaccharide may be released into blood in the course of IFD, including infection by species of Aspergillus, Candida, Fusarium, Trichosporin, Saccharomyces, Acremonium, and P. jiroveci. BG does not appear in blood from patients with infection by Cryptococcus spp. or the Mucorales. The assay for BG is not an immunoassay. Rather, assay for BG is based on the ability of the polysaccharide to activate factor G of the horseshoe crab coagulation cascade. With use of a chromogenic substrate, the test can detect BG levels as low as 1 pg/mL (Obayashi et al. 1995).
Assay for BG has value as a screen for presumptive diagnosis of invasive fungal infection (IFI). Early diagnosis allows for earlier initiation of antifungal therapy. Clinical studies have found BG testing to have a high sensitivity for diagnosis of IFI. The test has a strong negative predictive value, allowing the test to be used to exclude IFI. There are a number of limitations in the use of BG assay for diagnosis of IFI. First, BG is ubiquitous in the environment, which may produce false positive reactions. All testing materials must be glucan-free. Second, BG testing is typically performed at reference laboratories, which reduces time to result and discourages routine, potentially prospective, testing by clinicians. Finally, BG testing cannot be used to detect mucormycosis or cryptococcosis.
Strengths and Weaknesses of Antigen Testing
A particular advantage of testing for antigen is the possibility that antigen can be shed from a local site of infection to a body fluid such as blood or urine. As a consequence, it is possible to avoid highly invasive sample collection. Circulating or urinary antigen functions as a surrogate for the actual presence of the microbe. Second, it is possible to use antigen-detection platforms that can be inexpensive, rapid, and capable of use by personnel with limited training. This is the case with the CrAg LFA for diagnosis of cryptococcosis. Third, by judicious selection of antibodies, it is possible to produce a test with a broad or a very limited specificity, depending on the clinical needs for testing. A major limitation for antigen testing is the need to identify antigen surrogates for infection. CrAg is a well-characterized surrogate for infection; however, identification of similar surrogates for other infections is needed.
CONCLUDING REMARKS
Desirable properties of a diagnostic test are listed in Table 2. It is likely that several diagnostic platforms will be needed to meet the diverse requirements of different fungal infections and the resources available for testing. The ideal test would detect infection early in the course of disease, perhaps before the advent of symptoms. Early diagnosis would enable administration of antifungals at a time when treatment is most likely to be effective. For example, there is a 20% increase in mortality of invasive candidiasis if therapy is delayed by >12 h (Morrell et al. 2005). An example of a diagnostic test that can identify subacute infection is detection of cryptococcal polysaccharide in sera of patients who enter antiretroviral therapy who do not have symptoms of cryptococcal meningitis (Jarvis et al. 2009). Second, a test with strong negative predictive value would be of great value in managing patients at high risk for IFD. The neutropenic patient with fever of unknown origin that is unresponsive to broad-spectrum antibiotics is often treated empirically with antifungal agents. A test with a high negative predictive value could identify patients who should not be given antifungal therapy. This would reduce the cost of patient care and reduce development of antibiotic resistance.
Table 2.
Properties of ideal next-generation fungal diagnostics
| Detects infection early in course of disease |
| Strong negative predictive value |
| Short time to result; ideally near point of care |
| Low cost; inexpensive equipment/instrumentation |
| Use of noninvasive sample |
| Suitable for use by personnel with limited training |
| Minimal number of steps |
| Uncomplicated interpretation; no downstream analysis required |
Many laboratory tests for diagnosis of fungal infection are typically performed at reference laboratories, for example, tests for galactomannan or BG. The time to result presents a barrier to routine use of such tests. Similarly, high cost presents barriers to routine use of advanced testing for fungal infection. A major barrier to use of many diagnostic approaches is the need for invasive procedures for sample collection, particularly for high-risk patients. The ideal test would use readily accessible samples such as blood or urine. An example is the CrAg LFA, which has a high sensitivity for diagnosis of cryptococcosis using serum. This avoids the need for the lumbar puncture, a major advantage in resource-limited settings (Jarvis et al. 2013). Similarly, testing of urine for Histoplasma galactomannan enables diagnosis of disseminated histoplasmosis and is useful in early diagnosis of acute pulmonary histoplasmosis and in treatment follow-up (Wheat 2006). Finally, most diagnostic tests for IFI require a high level of operator expertise. Once again, this need for a relatively advanced infrastructure reduces access and raises barriers to testing.
New generations of fungal diagnostics must reconcile the diverse needs of patients in developed countries with advanced levels of medical care and those of patients in resource-limited settings. Many of the IFIs in countries with advanced medical care are a consequence of the ability to provide long-term critical care to patients or the use of highly immunosuppressive agents. In contrast, most deaths owing to fungal infection globally occur in patients with AIDS, most often in settings with little or no infrastructure. On a global level, fungal diagnostics must be able to function in a setting of limited infrastructure.
Is it possible to reconcile the needs of resource-limited countries and those of patients in settings of advanced medical care? Diagnostic tests that meet the criteria outlined in Table 2 may be able to do so. Clearly, low-cost testing that can be performed at the point-of-patient care is critical in resource-limited countries. However, these same tests may be able to dramatically improve outcome for the critically ill patient in advanced medical settings. For example, if testing could be performed at low cost with a noninvasive sample and produce a rapid result, such testing would enable prospective monitoring of those patients at high risk for IFD. Prospective monitoring would identify infection before the occurrence of overt symptoms and enable early treatment when it is most likely to be effective.
ACKNOWLEDGMENTS
This work is supported in part by National Institutes of Health (NIH) grants AI014209, AI085548, AI096945, and AI102311 to T.R.K. B.W. is supported by grant W81XWH-13-C-0103 from the U.S. Army Medical Research and Materiel Command, Office of Congressionally Directed Medical Research Programs, Joint Warfighter Medical Research Program.
Footnotes
Editors: Arturo Casadevall, Aaron P. Mitchell, Judith Berman, Kyung J. Kwon-Chung, John R. Perfect, and Joseph Heitman
Additional Perspectives on Human Fungal Pathogens available at www.perspectivesinmedicine.org
REFERENCES
- Abe A, Asano K, Sone T 2010. A molecular phylogeny-based taxonomy of the genus Rhizopus. Biosci Biotech Biochem 74: 1325–1331 [DOI] [PubMed] [Google Scholar]
- Ajello L, Hay RJ, ed. 1998. Topley and Wilson’s microbiology and microbial infections. Arnold, London [Google Scholar]
- Andrew M, Peever TL, Pryor BM 2009. An expanded multilocus phylogeny does not resolve morphological species within the small-spored Alternaria species complex. Mycologia 101: 95–109 [DOI] [PubMed] [Google Scholar]
- Azuma I, Kanetsuna F, Tanaka Y, Yamamura Y, Carbonell LM 1974. Chemical and immunological properties of galactomannans obtained from Histoplasma duboisii, Histoplasma capsulatum, Paracoccidioides brasiliensis and Blastomyces dermatitidis. Mycopathol Mycol Appl 54: 111–125 [DOI] [PubMed] [Google Scholar]
- Bader O 2013. MALDI-TOF-MS-based species identification and typing approaches in medical mycology. Proteomics 13: 788–799 [DOI] [PubMed] [Google Scholar]
- Barnes RA 2008. Early diagnosis of fungal infection in immunocompromised patients. J Antimicrob Chemother 61 (Suppl 1): i3–i6 [DOI] [PubMed] [Google Scholar]
- Bennett JE, Kwon-Chung KJ, Howard DH 1977. Epidemiologic differences among serotypes of Cryptococcus neoformans. Am J Epidemiol 105: 582–586 [DOI] [PubMed] [Google Scholar]
- Berbee ML, Pirseyedi M, Hubbard S 1999. Cochliobolus phylogenetics and the origin of known, highly virulent pathogens, inferred from ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycopath 91: 964–977 [Google Scholar]
- Bidartondo MI 2008. Preserving accuracy in GenBank. Science 319: 1616. [DOI] [PubMed] [Google Scholar]
- Bloomfield N, Gordon MA, Elmendorf DF Jr 1963. Detection of Cryptococcus neoformans antigen in body fluids by latex particle agglutination. Proc Soc Exp Biol Med 114: 64–67 [DOI] [PubMed] [Google Scholar]
- Carbone I, Kohn LM 1999. A method for designing primer sets for speciation studies in filamentous ascomycetes. Mycologia 91: 553–556 [Google Scholar]
- Chapman SW, Sullivan DC 2010. Blastomyces dermatitidis. In Principles and practice of infectious diseases (ed. Mandell GL, Bennett JE, Dolin R), pp. 3319–3332 Churchill Livingstone, Philadelphia [Google Scholar]
- Collier L, Balows A, Sussman M ed. 1998. Microbiology and microbial infections. Arnold, London [Google Scholar]
- Connolly PA, Durkin MM, Lemonte AM, Hackett EJ, Wheat LJ 2007. Detection of histoplasma antigen by a quantitative enzyme immunoassay. Clin Vaccine Immunol 14: 1587–1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly P, Hage CA, Bariola JR, Bensadoun E, Rodgers M, Bradsher RW, Wheat LJ 2012. Blastomyces dermatitidis antigen detection by quantitative enzyme immunoassay. Clin Vaccine Immunol 19: 53–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxatto A, Prod’hom G, Greub G 2012. Applications of MALDI-TOF mass spectrometry in clinical diagnostic microbiology. FEMS Microbiol Rev 36: 380–407 [DOI] [PubMed] [Google Scholar]
- Dannaoui E, Schwarz P, Slany M, Loeffler J, Jorde AT, Cuenca-Estrella M, Hauser PM, Shrief R, Huerre M, Freiberger T, et al. 2010. Molecular detection and identification of zygomycetes species from paraffin-embedded tissues in a murine model of disseminated zygomycosis: A collaborative European Society of Clinical Microbiology and Infectious Diseases (ESCMID) Fungal Infection Study Group (EFISG) evaluation. J Clin Microbiol 48: 2043–2046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deepe GS 2010. Histoplasma capsulatum. In Principles and practice of infectious diseases (ed. Mandell GL, Bennett JE, Dolin R), pp. 3305–3318 Churchill Livingstone, Philadephia [Google Scholar]
- de Hoog GS, Guarro J, Gene J, Figueras MJ 2000. Atlas of clinical fungi. Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands [Google Scholar]
- Dromer F, Mathoulin S, Dupont B, Letenneur L, Ronin O 1996. Individual and environmental factors associated with infection due to Cryptococcus neoformans serotype D. French Cryptococcosis Study Group. Clin Infect Dis 23: 91–96 [DOI] [PubMed] [Google Scholar]
- Durkin MM, Connolly PA, Wheat LJ 1997. Comparison of radioimmunoassay and enzyme-linked immunoassay methods for detection of Histoplasma capsulatum var. capsulatum antigen. J Clin Microbiol 35: 2252–2255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durkin M, Connolly P, Kuberski T, Myers R, Kubak BM, Bruckner D, Pegues D, Wheat LJ 2008. Diagnosis of coccidioidomycosis with use of the Coccidioides antigen enzyme immunoassay. Clin Infect Dis 47: e69–e73 [DOI] [PubMed] [Google Scholar]
- Ellepola ANB, Morrison CJ 2005. Laboratory diagnosis of invasive candidiasis. J Microbiol 43: 65–84 [PubMed] [Google Scholar]
- Fraser VJ, Jones M, Dunkel J, Storfer S, Medoff G, Dunagan WC 1992. Candidemia in a tertiary care hospital: Epidemiology, risk factors, and predictors of mortality. Clin Infect Dis 15: 414–421 [DOI] [PubMed] [Google Scholar]
- Galgiani JN 2010. Coccidioides species. In Principles and practice of infectious diseases (ed. Mandell GL, Bennett JE, Dolin R), pp. 3333–3344 Churchill Livingstone, Philadelphia [Google Scholar]
- Gao J, Takashima A 2004. Cloning and characterization of Trichophyton rubrum genes encoding actin, Tri r2, and Tri r4. J Clin Microbiol 42: 3298–3299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates-Hollingsworth MA, Kozel TR 2013. Serotype sensitivity of a lateral flow immunoassay for cryptococcal antigen. Clin Vaccine Immunol 20: 634–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass NL, Donaldson GC 1995. Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. Appl Environ Microbiol 61: 1323–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson RA, Seifert KA, Taylor JW, Wingfield MJ, Aime C, Asan A, et al. 2011. The Amsterdam declaration on fungal nomenclature. IMA Fungus 2: 105–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbett DS, Taylor JW 2013. Fungal systematics: Is a new age of enlightenment at hand? Nat Rev Microbiol 11: 129–133 [DOI] [PubMed] [Google Scholar]
- Hong SB, Cho HS, Shin HD, Frisvad JC, Samson RA 2006. Novel Neosartorya species isolated from soil in Korea. Int J Syst Evol Microbiol 56: 477–486 [DOI] [PubMed] [Google Scholar]
- Houbraken J, Samson RA 2011. Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Mycology 70: 1–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YT, Hung CC, Liao CH, Sun HY, Chang SC, Chen YC 2007. Detection of circulating galactomannan in serum samples for diagnosis of Penicillium marneffei infection and cryptococcosis among patients infected with human immunodeficiency virus. J Clin Microbiol 45: 2858–2862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis JN, Lawn SD, Vogt M, Bangani N, Wood R, Harrison TS 2009. Screening for cryptococcal antigenemia in patients accessing an antiretroviral treatment program in South Africa. Clin Infect Dis 48: 856–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis JN, Harrison TS, Lawn SD, Meintjes G, Wood R, Cleary S 2013. Cost effectiveness of cryptococcal antigen screening as a strategy to prevent HIV-associated cryptococcal meningitis in South Africa. PLoS ONE 8: e69288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karstaedt AS, Crewe-Brown HH, Dromer F 2002. Cryptococcal meningitis caused by Cryptococcus neoformans var gattii, serotype C, in AIDS patients in Soweto, South Africa. Med Mycol 40: 7–11 [DOI] [PubMed] [Google Scholar]
- Kontoyiannis DP, Sumoza D, Tarrand J, Bodey GP, Storey R, Raad II 2000. Significance of aspergillemia in patients with cancer: A 10-year study. Clin Infect Dis 31: 188–189 [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ 1997. Identification of clinically important ascomycetous yeasts based on nucleotide divergence in the 5′ end of the large-subunit (26S) ribosomal DNA gene. J Clin Microbiol 35: 1216–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE 1984a. Epidemiologic differences between the two varieties of Cryptococcus neoformans. Am J Epidemiol 120: 123–130 [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Bennett JE 1984b. High prevalence of Cryptococcus neoformans var gattii in tropical and subtropical regions. Zbl Bakt Hyg 257: 213–218 [PubMed] [Google Scholar]
- Lasker BA 2006. Nucleotide sequence-based analysis for determining the molecular epidemiology of Penicillium marneffei. J Clin Microbiol 44: 3145–3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann PF, Reiss E 1978. Invasive aspergillosis: Antiserum for circulating antigen produced after immunization with serum from infected rabbits. Infect Immun 20: 570–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley MD, Warnock DW, Morrison CJ 2006. Serological and molecular diagnosis of fungal infection. In Manual of molecular and clinical laboratory immunology (ed. Rose NR, Hamilton RG, Detrick B), pp. 569–605 American Society for Microbiology, Washington, D.C [Google Scholar]
- Litvintseva AP, Thakur R, Reller LB, Mitchell TG 2005. Prevalence of clinical isolates of Cryptococcus gattii serotype C among patients with AIDS in Sub-Saharan Africa. J Infect Dis 192: 888–892 [DOI] [PubMed] [Google Scholar]
- Litvintseva AP, Xu J, Mitchell TG 2011. Population structure and ecology of Cryptococcus neoformans and Cryptococcus gattii. In Cryptococcus: From human pathogen to model yeast (ed. Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A), pp. 97–111 American Society for Microbiology, Washington, D.C [Google Scholar]
- Liu YJ, Whelen S, Hall BD 1999. Phylogenetic relationships among ascomycetes: Evidence from an RNA Polymerse II subunit. Mol Biol Evol 16: 1799–1808 [DOI] [PubMed] [Google Scholar]
- Machetti M, Feasi M, Mordini N, Van Lint MT, Bacigalupo A, Latgé JP, Sarfati J, Viscoli C 1998. Comparison of an enzyme immunoassay and a latex agglutination system for the diagnosis of invasive aspergillosis in bone marrow transplant recipients. Bone Marrow Transpl 21: 917–921 [DOI] [PubMed] [Google Scholar]
- Maertens J, Verhaegen J, Demuynck H, Brock P, Verhoef G, Vandenberghe P, Van Eldere J, Verbist L, Boogaerts M 1999. Autopsy-controlled prospective evaluation of serial screening for circulating galactomannan by a sandwich enzyme-linked immunosorbent assay for hematological patients at risk for invasive aspergillosis. J Clin Microbiol 37: 3223–3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maertens J, Verhaegen J, Lagrou K, Van Eldere J, Boogaerts M 2001. Screening for circulating galactomannan as a noninvasive diagnostic tool for invasive aspergillosis in prolonged neutropenic patients and stem cell transplantation recipients: A prospective validation. Blood 97: 1604–1610 [DOI] [PubMed] [Google Scholar]
- Marimon R, Gene J, Cano J, Trilles L, Dos Santos Lazera M, Guarro J 2006. Molecular phylogeny of Sporothrix schenckii. J Clin Microbiol 44: 3251–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrell M, Fraser VJ, Koller MJ 2005. Delaying empiric treatment of Candida bloodstream infection until positive blood culture results are obtained: A potential risk factor for mortality. Antimicrob Agents Chemother 49: 3640–3645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Yoshida M, Mori T, Goto H, Yasuoka A, Iwasaki H, Teshima H, Kohno S, Horiuchi A, Ito A 1995. Plasma (1→3)-β-d-glucan measurement in diagnosis of invasive deep mycosis and fungal febrile episodes. Lancet 345: 17–20 [DOI] [PubMed] [Google Scholar]
- O’Donnell K, Kistler HC, Cigelnik E, Ploetz RC 1998. Multiple evolutionary origins of the fungus causing Panama disease of banana: Concordant evidence from nuclear and mitochondrial gene genealogies. Proc Natl Acad Sci 95: 2044–2049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell K, Nirenberg HI, Aoki T, Cigelnik E 2000. A multigene phylogeny of the Gibberella fujikuroi species complex: Detection of additional phylogenetically distinct species. Mycoscience 41: 61–78 [Google Scholar]
- O’Donnell K, Lutzoni FM, Ward TJ, Benny GL 2001. Evolutionary relationships among mucoralean fungi (Zygomycota): Evidence for family polyphyly on a large scale. Mycologia 93: 286–297 [Google Scholar]
- Ostrosky-Zeichner L 2012. Invasive mycoses: Diagnostic challenges. Am J Med 125: S14–S24 [DOI] [PubMed] [Google Scholar]
- Ostrosky-Zeichner L, Pappas PG 2006. Invasive candidiasis in the intensive care unit. Crit Care Med 34: 857–863 [DOI] [PubMed] [Google Scholar]
- Peterson SW 2008. Phylogenetic analysis of Aspergillus species using DNA sequences from four loci. Mycologia 100: 205–206 [DOI] [PubMed] [Google Scholar]
- Petraitiene R, Petraitis V, Witt JR, Durkin MM, Bacher JD, Wheat LJ, Walsh TJ 2011. Galactomannan antigenemia after infusion of gluconate-containing Plasma-Lyte. J Clin Microbiol 49: 4330–4332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petti CA, Bosshard PP, Brandt ME, Clarridge JE, Feldblyum TV, Foxall P, Furtado MR, Pace N, Procop G 2008. Interpretive criteria for identification of bacteria and fungi by DNA target sequencing; approved guideline. In MM18-P. Clinical Laboratory Standards Institute, Wayne, PA [Google Scholar]
- Pinel C, Fricker-Hidalgo H, Lebeau B, Garban F, Hamidfar R, Ambroise-Thomas P, Grillot R 2003. Detection of circulating Aspergillus fumigatus galactomannan: Value and limits of the Platelia test for diagnosing invasive aspergillosis. J Clin Microbiol 41: 2184–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posteraro B, De Carolis E, Vella A, Sanguinetti M 2013. MALDI-TOF mass spectrometry in the clinical mycology laboratory: Identification of fungi and beyond. Expert Rev Proteomics 10: 151–164 [DOI] [PubMed] [Google Scholar]
- Preuner S, Lion T 2013. Species-specific identification of a wide range of clinically relevant fungal pathogens by the Luminex® xMAP technology. Methods Mol Biol 968: 119–139 [DOI] [PubMed] [Google Scholar]
- Reiss E, Lehmann PF 1979. Galactomannan antigenemia in invasive aspergillosis. Infect Immun 25: 357–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson RA, Yilmaz N, Houbraken J, Spierenburg H, Seifert KA, Peterson SW, Varga J, Frisvad JC 2011. Phylogeny and nomenclature of the genus Talaromyces and taxa accommodated in Penicillium subgenus Biverticillium. Stud Mycol 70: 159–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saubolle MA, McKellar PP, Sussland D 2007. Epidemiologic, clinical, and diagnostic aspects of coccidioidomycosis. J Clin Microbiol 45: 26–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach WJ, Mitchell TG, Schell WA, Espinel-Ingroff A, Coico RF, Walsh TJ, Perfect JR 2003. Status of medical mycology education. Med Mycol 41: 457–467 [DOI] [PubMed] [Google Scholar]
- Stynen D, Sarfati J, Goris A, Prévost MC, Lesourd M, Kamphuis H, Darras V, Latgé JP 1992. Rat monoclonal antibodies against Aspergillus galactomannan. Infect Immun 60: 2237–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stynen D, Goris A, Sarfati J, Latgé JP 1995. A new sensitive sandwich enzyme-linked immunosorbent assay to detect galactofuran in patients with invasive aspergillosis. J Clin Microbiol 33: 497–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita T, Nakajima M, Ikeda R, Matsushima T, Shinoda T 2002. Sequence analysis of the ribosomal DNA intergenic spacer 1 regions of Trichosporon species. J Clin Microbiol 40: 1826–1830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugui JA, Peterson SW, Clark LP, Nardone G, Folio L, Riedlinger G, Zerbe CS, Shea Y, Henderson CM, Zelazny AM, et al. 2012. Aspergillus tanneri sp. nov., a new pathogen that causes invasive disease refractory to antifungal therapy. J Clin Microbiol 50: 3309–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulahian A, Boutboul F, Ribaud P, Leblanc T, Lacroix C, Derouin F 2001. Value of antigen detection using an enzyme immunoassay in the diagnosis and prediction of invasive aspergillosis in two adult and pediatric hematology units during a 4-year prospective study. Cancer 91: 311–318 [DOI] [PubMed] [Google Scholar]
- Thakur R, Steele KT, Nthobatsang R, Bafana M, Steenhoff AP, Bisson GP 2009. Prevalence of C. neoformans and C. gattii among patients with cryptococcal meningitis at a tertiary hospital in Gaborone, Botswana: March 2005–February 2007, Abstract 1048. 47th Annual Meeting IDSA, Philadelphia [Google Scholar]
- Tobón AM, Agudelo CA, Rosero DS, Ochoa JE, De Bedout C, Zuluaga A, Arango M, Cano LE, Sampedro J, Restrepo A 2005. Disseminated histoplasmosis: A comparative study between patients with acquired immunodeficiency syndrome and non-human immunodeficiency virus-infected individuals. Am J Trop Med Hyg 73: 576–582 [PubMed] [Google Scholar]
- Wheat LJ 2006. Improvements in diagnosis of histoplasmosis. Expert Opin Biol Ther 6: 1207–1221 [DOI] [PubMed] [Google Scholar]
- Wheat LJ, Kohler RB, Tewari RP 1986. Diagnosis of disseminated histoplasmosis by detection of Histoplasma capsulatum antigen in serum and urine specimens. N Engl J Med 314: 88. [DOI] [PubMed] [Google Scholar]
- Wheat J, Wheat H, Connolly P, Kleiman K, Supparatpinyo K, Nelson K, Bradsher R, Restrepo A 1997. Cross-reactivity in Histoplasma capsulatum variety capsulatum antigen assays of urine samples from patients with endemic mycoses. Clin Infect Dis 24: 1169–1171 [DOI] [PubMed] [Google Scholar]
- Wheat LJ, Garringer T, Brizendine E, Connolly P 2002. Diagnosis of histoplasmosis by antigen detection based upon experience at the histoplasmosis reference laboratory. Diag Microbiol Infect Dis 43: 29–37 [DOI] [PubMed] [Google Scholar]
- Wheat LJ, Hackett E, Durkin M, Connolly P, Petraitiene R, Walsh TJ, Knox K, Hage C 2007. Histoplasmosis-associated cross-reactivity in the BioRad Platelia Aspergillus enzyme immunoassay. Clin Vaccine Immunol 14: 638–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TJ, Bruns TD, Lee SB, Taylor JW 1990. Amplification and sequencing of fungal ribosomal RNA genes for phylogenetics. Academic, New York [Google Scholar]
- Williams B, Fojtasek M, Connolly-Strigfield P, Wheat J 1994. Diagnosis of histoplasmosis by antigen detection during an outbreak in Indianapolis, Ind. Arch Pathol Lab Med 118: 1205–1208 [PubMed] [Google Scholar]
- World Health Organization Cryptococcal Working Group 2011. Rapid advice: Diagnosis, prevention and management of cryptococcal disease in HIV-infected adults, adolescents and children. World Health Organization, Geneva: [PubMed] [Google Scholar]
- Xavier MO, Pasqualotto AC, Cardoso IC, Severo LC 2009. Cross-reactivity of Paracoccidioides brasiliensis, Histoplasma capsulatum, and Cryptococcus species in the commercial Platelia Aspergillus enzyme immunoassay. Clin Vaccine Immunol 16: 132–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz N, Houbraken J, Hoekstra ES, Frisvad JC, Visagie CM, Samson RA 2012. Delimitation and characterisation of Talaromyces purpurogenus and related species. Persoonia 29: 39–54 [DOI] [PMC free article] [PubMed] [Google Scholar]


