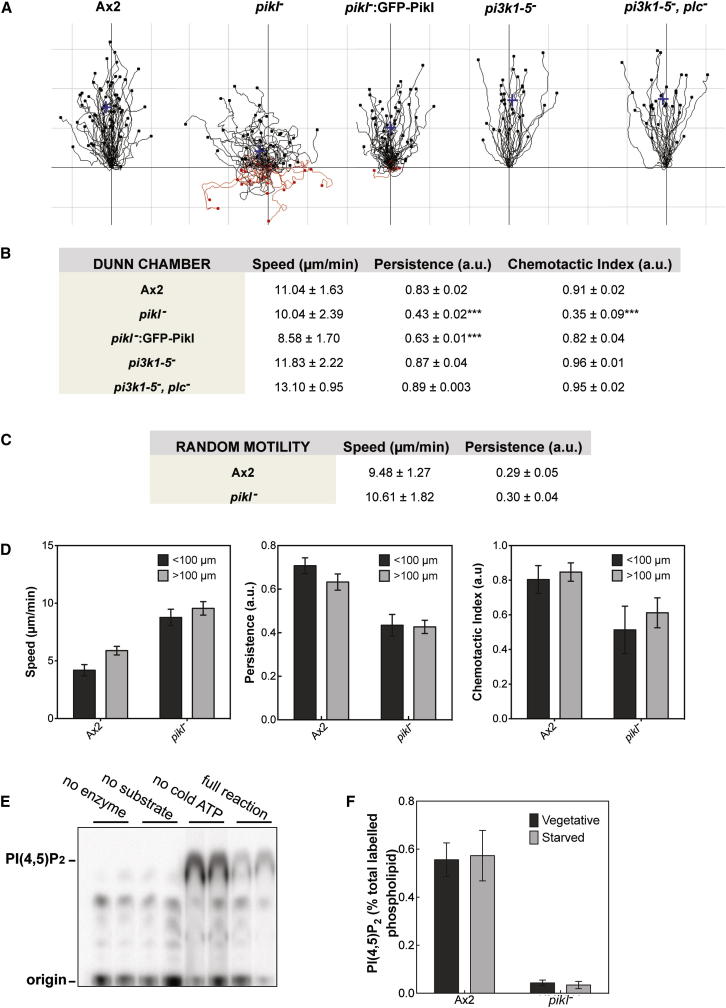
Figure 1.
Loss of the Active PIP5K, PikI, Severely Impairs Gradient Sensing
(A) Representative tracks of cells (strains indicated) chemotaxing toward cAMP in a Dunn chemotaxis chamber. Red tracks represent cells that have moved downgradient overall. Blue crosses represent the center of mass of the tracks. Grid lines are 100 μm apart.
(B) Table of chemotactic parameters obtained by tracking cells in a Dunn chemotaxis chamber. Speed is defined as accumulated distance divided by time, persistence as the Euclidean distance divided by accumulated distance, and chemotactic index as the cosine of the angle between the net distance traveled in the direction of the gradient and the Euclidean distance. Data represent the grand mean ± SD and were calculated from mean values obtained on at least two different days (Ax2: 6 days, n = 350 cells; pikI−: 3 days, n = 173 cells; pikI−: GFP-PikI: 3 days, n = 173 cells; pi3k1−5−: 4 days, n = 120; pi3k1−5−,plc−: 2 days, n = 60 cells. Means for each day were compared using a one-way ANOVA with Tukey’s post hoc test. ∗∗∗p < 0.001, mutant and rescued strains versus wild-type.
(C) Table of vegetative random motility parameters. Data represent the grand mean ± SD and were calculated from mean values obtained on three different days (Ax2 and pikI−, n = 60).
(D) Graphs representing speed, persistence, and chemotactic index of cells moving toward a micropipette filled with 10 μM cAMP (strong gradient). The chemotactic abilities of cells closer to the micropipette (<100 μm) were compared with those further from it (>100 μm). Data represent the mean ± SEM from three different days (Ax2 < 100 μm, n = 14; Ax2 > 100 μm, n = 45; pikI− < 100 μm, n = 17; pikI− > 100 μm, n = 42).
(E) Thin-layer chromatography separation of products from a PIP kinase assay, showing that recombinant PikI is an active PIP5K.
(F) Quantification of PI(4,5)P2 levels in cells labeled with 32Pi over several generations to reach steady-state labeling. PI(4,5)P2 levels in both vegetative and 5 hr-starved cells were compared. Data represent mean ± SEM calculated from at least three independent experiments.