Figure 4.
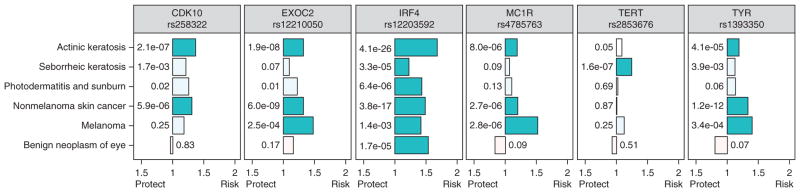
Risk variants for skin phenotypes have different pleiotropy patterns. Association odds ratios are graphed on the x axis and P-values (numbers next to the bars) are from the PheWAS analysis for that SNP. All SNPs use the minor allele as the coded allele, except rs2853676 (TERT). Darker colored bars represent significant associations, calculated as P = 0.05 divided by the number of associations displayed, or 0.05/(6 phenotypes*6 SNPs) = 1.4 × 10−3. Tests for heterogeneity revealed significant heterogeneity among the six phenotypes (I2 = 59–94%, all P < 0.05) and among the six SNPs (I2 = 23–83%, all P < 0.05). Bars oriented leftward toward “protect” represent SNPs in which the coded allele favors decreased prevalence of disease, and bars oriented rightward toward “risk” represent coded alleles favoring increased prevalence of disease.
