SUMMARY
Condensin is a central regulator of mitotic genome structure, with mutants showing poorly condensed chromosomes and profound segregation defects. Here we identify NCT complex, comprising the Nrc1 BET-family tandem bromodomain protein (SPAC631.02), Casein Kinase II (CKII) and several TAFs, as a regulator of condensin function. We show that NCT and condensin bind similar genomic regions, but only briefly co-localize during the periods of chromosome condensation and decondensation. This pattern of NCT binding at the core centromere, the region of maximal condensin enrichment, tracks the abundance of acetylated histone H4, as regulated by the Hat1-Mis16 acetyltransferase complex and recognized by the first Nrc1 bromodomain. Strikingly, mutants in NCT or Hat1-Mis16 restore the formation of segregation-competent chromosomes in cells containing defective condensin. These results are consistent with a model where NCT targets CKII to chromatin in a cell cycle-directed manner to modulate the activity of condensin during chromosome condensation and decondensation.
Keywords: Bromodomain, Casein Kinase II, condensin, histone acetylation, mitosis
INTRODUCTION
The genome has to be faithfully transmitted to each daughter at cell division. To this end the interphase chromatin is condensed to individual chromosomes at early mitosis, providing the structure needed to survive sister-chromatid separation. A major regulator of this extensive remodeling is the pentameric condensin complex, comprising two SMC (Structural Maintenance of Chromosomes) ATPases, a kleisin, and two HEAT-repeat proteins (Cuylen and Haering, 2011; Hirano, 2012; Onn et al., 2007; Wood et al., 2010). Cells containing mutants in each condensin subunit show poorly structured mitotic chromosomes and profound segregation defects, including the fission yeast (Schizosaccharomyces pombe; Sp) cut+ phenotype where the division septum cuts through unsegregated chromosomes at the metaphase plate (Saka et al., 1994).
Many metazoans contain two condensin complexes (I and II) that pair the same SMCs with alternative accessory subunits. This allows each complex to function independently, such that vertebrate condensin II regulates early chromosome condensation in prophase, and condensin I then loads in prometaphase to complete the reaction (Hirota et al., 2004; Ono et al., 2004; Ono et al., 2003). Fission yeast, in contrast, relies on a single condensin I that is presumed to regulate chromosome condensation through mitosis. The precise means by which any condensin regulates chromosome structure is unclear. In vitro analyses show that the immunopurified complex can introduce positive supercoils to relaxed circular DNA (in concert with topoisomerase I) and induce chiral knotting in nicked DNA (with topoisomerase II) (Kimura et al., 1999). During in vivo condensation condesin is also thought to generate higher order structures by directly linking distant regions of a chromosome fiber (Cuylen and Haering, 2011; Hirano, 2012; Wood et al., 2010).
Condensin is regulated by multiple means at various cell-cycle stages, including differential compartmentalization, chromosomal association and covalent modification. In this manner fission yeast condensin localizes to the cytoplasm for much of the cell cycle but is phosphorylated by Cdc2 at early mitosis and transported into the nucleus for loading to the centromere, rDNA and specific locations along the chromosome arms (Nakazawa et al., 2008; Sutani et al., 1999). This preference for a range of genomic features is likely mediated by binding of the condensin subunits to various chromatin marks (e.g. H4-K20Me1 and the H2A/H2A.Z N-terminal tails (Liu et al., 2010; Tada et al., 2011)) and adaptor proteins (e.g. Csm1/Lrs4, Scc2/Scc4, TFIIIB/TFIIIC, Cti1, Cti2, PARP, PP2A, AKAP95 and Pku80 (Chen et al., 2004; D’Ambrosio et al., 2008; Heale et al., 2006; Johzuka and Horiuchi, 2009; Steen et al., 2000; Takemoto et al., 2009; Tanaka et al., 2012)).
Chromosome condensation is unlikely a simple direct consequence of condensin - DNA binding: the complex also has to be activated. Covalent modification is presumed central to this regulation, with many of the condensin subunits extensively phosphorylated, acetylated and sumoylated (Bazile et al., 2010; Choudhary et al., 2009; Cuylen and Haering, 2011; Hirano, 2012). Phosphorylation is the most studied, where distinct events can inhibit (if catalyzed by Casein Kinase II (CKII)) or activate (if catalyzed by various mitotic kinases) condensins’ in vitro supercoiling activity (Bazile et al., 2010). In a related fashion the human hsp90, hsp27, hsp70 and c-fos genes recruit PP2A phosphatase to accelerate their re-expression in the following G1 by dephosphorylating / inactivating any co-localized condensin (Sarge and Park-Sarge, 2009; Xing et al., 2008).
In this study we identify novel regulators of fission yeast condensin, and thus mitotic chromosome function. We describe the Hat1-Mis16 acetyltransferase complex, show that this contributes to the acetylation of histones H3 and H4 at the core centromere (the region of peak condensin loading), and demonstrate that these modifications are cell-cycle regulated and anti-correlated with condensin binding through mitosis. We also describe the NCT complex, comprising the Nrc1 bromodomain (SPAC631.02), CKII and several TAF proteins, and show that NCT and condensin bind similar genomic regions, but only briefly co-localize during the periods of chromosome condensation and decondensation. Importantly we find that mutants in Hat1-Mis16 or NCT restore the formation of segregation-competent chromosomes in cells containing defective condensin. Our findings are consistent with a model where NCT targets CKII to chromatin in a cell cycle-directed manner to modulate condensins’ catalytic activity, thus regulating chromosome condensation and decondensation.
RESULTS
Deletions of the Hat1 acetyltransferase and Nrc1 bromodomain rescue the anaphase segregation defects of mutant condensin
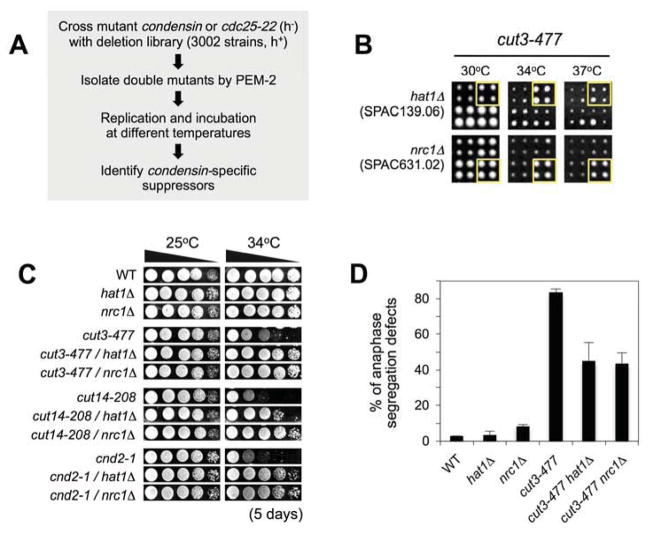
To begin this study we used a genetics approach to identify second site deletions that specifically rescue the lethality of conditional condensin mutants (the temperature sensitive (ts) alleles cut3-477 or cut14-208), expecting this would uncover novel regulators of mitotic chromosome function (Figure 1A). Screening 3,002 fission yeast deletions (~ 75% of the non-essential genome (Kim et al., 2010)) identified suppressive deletions of two factors with likely roles in the regulation or recognition of acetyl-lysine (Kac): Hat1 acetyltransferase and SPAC631.02 (hereafter called Nrc1; Negative Regulator of Condensin 1), a poorly characterized protein containing a BET-family tandem bromodomain (Figures 1B & S1A). Direct testing confirmed that hat1Δ and nrc1Δ also suppressed a ts allele of the condensin kleisin subunit (cnd2-1) (Figure 1C). Furthermore each deletion significantly improved the anaphase chromosome structure achievable by cut3-477 alone (Figure 1D), indicating that their suppression of mutant condensin is mediated at mitosis.
Figure 1. Deletions of the Hat1 acetyltransferase and Nrc1 bromodomain rescue mutant condensin.
(A) The PEM-2 approach was used to place conditional temperature sensitive (ts) alleles of condensin (cut3-477, cut14-208) or cdc25 phosphatase (cdc25-22) in the context of deletions of ~ 75% of the non-essential Sp genome. (B) condensin-specific suppressors include individual deletions of Hat1 acetyltransferase and the Nrc1 bromodomain. Double-mutant arrays were pinned in quadruplicate and incubated as indicated (mutants of interest are boxed; panel are size-standardized to facilitate cross-comparison). (C) hat1Δ and nrc1Δ rescue condensin ts alleles (cut3-477, cut14-208 and cnd2-1) in direct testing. Strains were isolated by crossing and tetrad dissection, spotted as 10-fold serial dilutions onto non-selective YES media, and incubated as indicated. WT, wild type. (D) hat1Δ and nrc1Δrepair the anaphase chromosome segregation defects of cut3-477. Replicate cultures were shifted from 25°C to 34°C (non-permissive for cut3-477) for three hours, fixed, and the percentage (mean ± s.d.) of late anaphase cells with chromosome segregation defects determined.
Hat1-Mis16 regulates the core centromeric histone acetylation that anti-correlates with condensin occupancy through mitosis
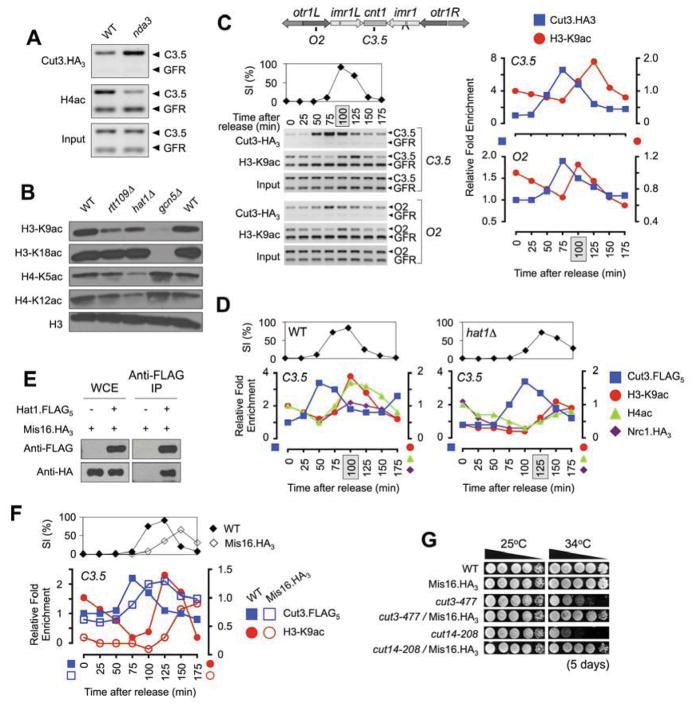
The identification of hat1Δ and nrc1Δ as condensin suppressors suggested that specific acetyl-lysines might play a role in condensin function. To investigate this we first examined the core centromere, the region of maximal condensin enrichment at mitosis (Nakazawa et al., 2008) (Figure S1B–D). Of note H4ac at this location is dramatically reduced in a metaphase - arrested population when condensin is maximally loaded (Figure 2A). Hat1 contributes to the acetylation of histones H3 and H4 (Benson et al., 2007) (Figure 2B), so we used Chromatin ImmunoPrecipitation (ChIP) to monitor these modifications in cell-cycle synchronized populations. In this approach H3ac and H4ac levels at the core and outer centromere temporally anti-correlated with condensin: i.e. they reduced as Cut3 bound in prometaphase and increased as Cut3 departed in G1/S (Figures 2C & S1E–I). Hat1 contributed to this acetylation profile, since hat1Δ constitutively reduced centromeric H3ac and H4ac levels and delayed mitotic progression and condensin binding (Figures 2D & S1I). These findings identified Hat1 acetyltransferase as a potential regulator of mitosis and centromere function.
Figure 2. Centromeric H3ac and H4ac levels anti-correlate with condensin through mitosis in a Hat1-dependent manner.
(A) H4ac is reduced at the core centromere in metaphase-arrested cells. WT or nda3-KM311 cells were placed at a restrictive temperature for the cold-sensitive tubulin allele (20°C, 6 h) to induce spindle-dependent metaphase arrest (Hiraoka et al., 1984) and ChIP used to monitor condensin (represented by the Cut3 subunit) and H4ac in each population (WT is asynchronous). In each duplex PCR the upper band is the test region (C3.5, core centromere: see Figure 2C), lower band (GFR, Gene Free Region) is a condensin-free location as a background control (Kim et al., 2009). Input tests primer efficiency in each sample. (B) Hat1 contributes to the acetylation of histones H3 and H4. Whole cell extracts (WCEs) were isolated from the indicated strains and the relative level of each acetylated species determined by immunoblotting. Total H3 is a loading control. (C) Centromeric H3-K9ac levels anti-correlate with condensin. Cells (additionally containing cdc25-22) were arrested at G2/M before synchronous release (as in Figure S1B) and ChIP used to monitor the indicated factors or histone modifications at the chromosome I centromere core or outermost repeats (C3.5 or O2; see upper schematic). The septation index (SI: peak shaded) and control ChIPs confirmed synchronous progression and equivalent antibody access at each time point (see also Figure S1E–J). The specific ChIP signal at each location / time point is expressed relative to the respective T0 (set to 1). (D) Nrc1 binding at the centromere core parallels Hat1-dependent H3-K9ac and H4ac, and anti-correlates with condensin (see also Figure S1I). Each ChIP signal is expressed relative to the respective WT T0 (set to 1). (E) Hat1 co-precipitates Mis16 (see also Table S1). Immunoblots of epitope tagged factors in WCEs or immunoprecipitates (IPs) are as indicated. (F) Mis16.HA3 binding at the centromere core parallels mitotically regulated H3-K9ac (see also Figure S1J). The timing of peak septation and condensin binding in Mis16.HA3 indicates delayed mitotic progression (as in hat1Δ: panel D). (G) Mis16.HA3 rescues condensin mutants (see also Figure S1K). Strains were spotted as 10-fold serial dilutions onto YES plates.
We next asked how Hat1 might itself be regulated, and sought any attendant proteins by sequential purification / LC-MS from whole cell extracts (WCEs) containing Hat1.TAP. This identified Mis16 (~ 37% identity to Hat2 of the budding yeast Hat1-Hat2 complex) as an associated protein, a relationship confirmed by co-immunoprecipitation (Figure 2E). Mis16 was previously identified in complex with Mis18 (Hayashi et al., 2004), but our reciprocal purifications from Mis16.TAP and Mis18.TAP WCEs distinguished two complexes: [Mis16 - Mis18 - SPBC27B12.02 - SPBC776.16] and [Hat1 - Mis16] (Table S1). Thus fission yeast likely contains at least two Mis16 complexes with distinct functions. The previously described [Mis16 - Mis18] regulates the centromeric loading of histone variant CenH3 (Sp Cnp1) via the Scm3 chaperone (Hayashi et al., 2004; Pidoux et al., 2009; Williams et al., 2009), while the newly identified [Hat1 - Mis16] may regulate histone acetylation and condensin function in a more general fashion. Supporting this proposal Mis18.GFP is exclusively centromeric, while Mis16.GFP shows diffuse nuclear fluorescence with centromeric enrichment (Hayashi et al., 2004).
Previous studies indicate that Mis16 dissociates from the centromere in prometaphase and returns at telophase / G1 (Hayashi et al., 2004), a pattern that would allow it to contribute to the mitotic regulation of H3ac and H4ac levels at this location (e.g. Figure 2C). To examine this further we subjected a strain containing epitope-tagged Mis16 to synchronous ChIP and observed that Mis16.HA3 indeed associated at the core centromere in a manner correlated with H3ac (Figure S1J). However these cells also showed mutant phenotypes reminiscent of hat1Δ, including reduced centromeric H3ac levels, delays in mitotic progression (compare Figures 2F & 2D), and the rescue of condensin mutants (Figures 2G & S1K). This may suggest that Mis16 mediates the association of Hat1 with chromatin, using its C-terminal WD40-repeats to target the acetyltransferase to specific locations (Ruthenburg et al., 2006; Trievel and Shilatifard, 2009): epitope-tagging at the Mis16 C-terminus could impede this function, and thus phenocopy hat1Δ.
The first bromodomain of Nrc1 binds H4ac
The temporal anti-correlation of H3ac/H4ac with condensin at the centromere (e.g. Figure 2D) raised the possibility that these modifications could influence the condensation / decondensation of this region. We further considered that this could be effector-mediated, such that the acetylations recruit a negative regulator of condensin.
The primary module with Kac-specificity is the ~ 110 amino-acid bromodomain (BD) (Filippakopoulos et al., 2012), so it was of interest that deletion of the tandem-BD protein Nrc1 also suppressed condensin mutants (Figure 1B–D). Indeed, there was a dose-dependent relationship between these factors, with reduced or increased Nrc1 levels respectively suppressing or enhancing the growth defects of mutant condensin (Figure S2A–C). Furthermore, genetic analyses of nrc1Δ identified negative interactions with mutants related to chromosome segregation, centromere identity, histone deacetylation, and condensin function (Roguev et al., 2008; Ryan et al., 2012) (Figure S2D). Importantly, the latter group included the deletion (pht1Δ) or unacetylatable mutation (pht1-NΔ, 4KR, or 4KQ) of histone variant H2A.Z, all of which show the premature dissociation of condensin from anaphase chromosomes (Kim et al., 2009). Together this collection of genetic interactions strongly supported a role for Nrc1 in mitotic chromosome function.
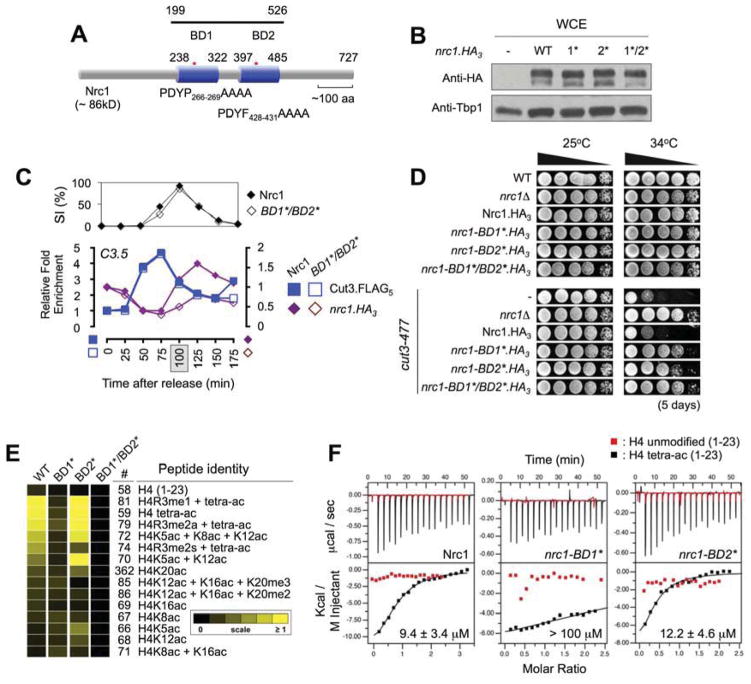
Various models of BDs in complex with specific Kac-peptides identify a conserved region comprising a left-handed bundle of four α-helices (αZ, A, B, C) linked by the ZA and BC loops that contribute to substrate specificity (Dhalluin et al., 1999; Jacobson et al., 2000; Owen et al., 2000). We used these structures to mutagenize the region containing an invariant tyrosine in the ZA loop of each Nrc1 BD (BD1*, nrc1-PDYP266–269AAAA; BD2*, nrc1-PDYF428–431AAAA; BD1*/BD2*: Figure 3A), which should significantly reduce their affinity for Kac (Dhalluin et al., 1999). Immunoblotting confirmed that each allele was expressed at a comparable level to wild type (WT, Figure 3B) but nrc1-BD1*/BD2* showed reduced centromeric recruitment after mitosis (Figure 3C) indicating that its bromodomains contribute to the association of Nrc1 with chromatin. This appears essential to Nrc1 function since each BD mutant also rescued mutant condensin (Figure 3D), recapitulating this phenotype of nrc1Δ (e.g. Figure 1C).
Figure 3. Nrc1-BD1 specifically interacts with Hat1-acetylated histone H4.
(A) Schematic indicates the relative location of both Nrc1 bromodomains, the BD1* and BD2* mutations, and the region cloned for recombinant expression. (B) Mutation of the αZA loop of each BD has no impact on Nrc1 protein stability. WCEs were isolated and the relative level of each nrc1.HA3 determined by immunoblotting. Tbp1 is a loading control. (C) Nrc1 requires its BDs for efficient recruitment to the centromere. Cells were synchronized and ChIPed as in Figure 2C, with the specific ChIP signal at each time point expressed relative to the respective WT T0 (set to 1). (D) Mutation of the Nrc1 BDs rescues cut3-477. Strains were spotted as 10-fold serial dilutions onto YES plates. (E) Nrc1-BD1 shows strong specificity for the hyperacetylated N-terminus of histone H4 (see also Figure S2E–G). Purified recombinants were used to probe histone-peptide arrays and areas of relative enrichment identified. (F) Nrc1-BD1 binds with μM affinity to H4tetra-ac but not to unacetylated H4 (both peptides: H4 residues 1–23). Time courses of raw injection heats (isotherm, upper panels) and normalized binding enthalpies are shown.
Individual BDs display selectivity for specific acetyl-lysines, most frequently (though unlikely exclusively) those on the histone N-termini (Dhalluin et al., 1999; Filippakopoulos et al., 2012; Owen et al., 2000). To examine such specificity for the Nrc1 BDs we purified recombinant WT and mutant forms of the domains (Figure 3A), and used each to probe a comprehensive array of histone peptides containing various combinations of post-translational modifications (Fuchs et al., 2010; Rothbart et al., 2012). In this approach the WT and BD2* recombinants (but not BD1* or BD1*/BD2*) showed strong specificity for peptides representing a hyper-acetylated form of the histone H4 N-terminus (Figures 3E & S2E–G). This was further investigated by isothermal titration calorimetry (ITC), where the WT and BD2* recombinants bound with ~ 10 μM affinity to an H4tetra-ac peptide, an ability not exhibited by BD1* (Figure 3F). Together, these analyses indicate that Nrc1 binds H4ac via BD1, a specificity that reflects the correlation between centromeric H4ac and Nrc1 levels through mitosis (Figure 2D). Our inability to distinguish Nrc1-BD2 binding in these in vitro studies does not indicate a lack of function, with the importance of this domain demonstrated by phenotypic testing (Figure 3C). It is possible that BD2 binds arrayed histone peptides weaker than the detection threshold, or recognizes an unrepresented histone or non-histone Kac-substrate.
Identification and characterization of the NCT complex
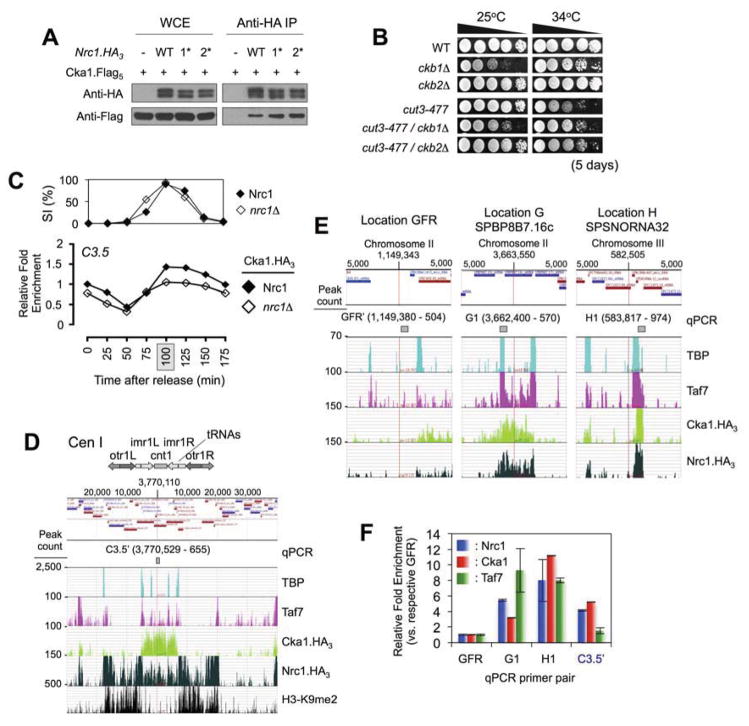
The above results suggest that Nrc1 could be an effector, recruited by specific mitotically regulated histone acetylations to regulate condensin function. However, other than its tandem bromodomains to mediate chromatin binding, Nrc1 contains no obvious functional domains (Figure 3A). To address this we generated an Nrc1.TAP strain to isolate any associated proteins by sequential purification / LC-MS. This identified all three subunits of the Casein Kinase II holoenzyme (CKII: Cka1, Ckb1 and Ckb2) and five TAFs (TBP associated factors) as binding partners (Figure 4A & Table S1). The association with CKII was of particular interest since the human form of this kinase hyper-phosphorylates four subunits of condensin I in interphase, inhibiting its supercoiling activity (Takemoto et al., 2006). This kinase-substrate relationship appears conserved across evolution, with proteomic analyses identifying phosphopeptides corresponding to consensus CKII substrates on the non-SMC subunits of budding yeast condensin (Bazile et al., 2010; Beltrao et al., 2009; Smolka et al., 2007). Fission yeast condensin contains > 100 consensus CKII sites (SXXE/D: high threshold predictions by GPS 2.1 online (Xue et al., 2008)) so we used a genetic approach to investigate any potential relationship between these factors. The CKII α-kinase subunit (Cka1) is essential for viability, but both β-regulatory subunits (Ckb1, Ckb2) are individually redundant (Roussou and Draetta, 1994) and ckb1Δ or ckb2Δ each rescued condensin mutants (Figure 4B), with ckb1Δ also improving the mitotic chromosome architecture achieved by cut3-477 alone (Figure S1K). This penetrant rescue of mutant condensin suggests a core role for the kinase, despite its multiple substrates (Filhol and Cochet, 2009). The relative strength of rescue (ckb1Δ > ckb2Δ) may reflect the differential contribution of each β-regulatory subunit to activity of the α-kinase in vivo (as suggested by cell growth, with cka1Δ lethal > ckb1Δ sick > ckb2Δ WT-like: Figure 4B).
Figure 4. Identification / initial characterization of the NCT (Nrc1 - CKII - TAFs) complex.
(A) Nrc1 (independent of its BDs) co-precipitates Cka1 (see also Table S1). Epitope tagged factors from WCEs or IPs were immunoblotted as indicated. (B) Deletion of the CKII regulatory β subunits rescues cut3-477 (ckb1Δ > ckb2Δ: see also Figure S1K). Strains were spotted as 10-fold serial dilutions onto YES plates. (C) Cka1 occupancy at the core centromere through mitosis is partially dependent on (and parallels: see Figure 3D) Nrc1. All ChIP signals are expressed relative to WT T0 (set to 1). (D) Nrc1, Cka1 and Taf7 enrichment across the chromosome 1 centromere (see also Figure S3). ChIP-seq data from asynchronous cells was loaded into Genplay as individual tracks relative to the annotated Sp genome (the online version of this paper links to a Genplay project to dynamically visualize each track). TBP identifies the tRNAs that demark the inner/outer centromere and pericentromeric boundary; H3-K9me2 identifies the heterochromatic outer repeats. Upper schematic depicts CEN1 structure; lower track genomic elements (e.g. genes or tRNAs) in the ~70 kb window region. (E) Nrc1, Cka1 and Taf7 co-localization at representative locations on the chromosome arms (see also Figure S3). Each 10 kb window is named for the centered genomic feature. GFR encompasses the condensin-free region used as a background control for ChIP (e.g. Figure 2C). (F) ChIP confirms ChIP-seq predictions of Nrc1, Cka1 and Taf7 enrichment at the chromosome arms (GFR′, G1, H1) and core centromere (c3.5′). Samples were prepared from asynchronous cells and analyzed by pPCR (rather than duplex PCR as previously) with primers flanking the regions indicated in panels D & E. The enrichment of each factor at each location is normalized to GFR (set to 1 for each IP).
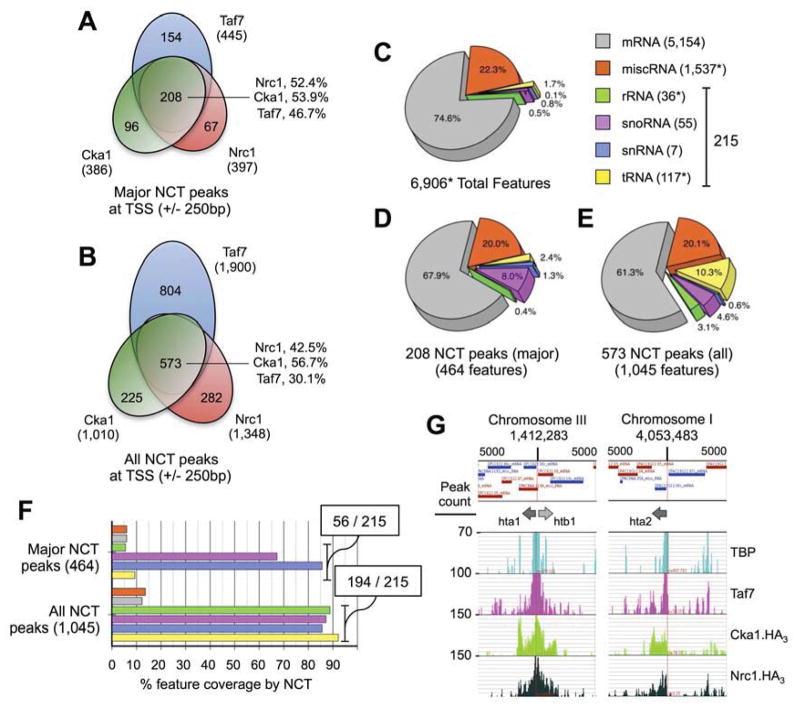
CKII is a promiscuous enzyme, so in vivo specificity is achieved by associated proteins targeting the kinase to distinct subcellular locations (Filhol and Cochet, 2009). In this manner Cka1 binding at the core centromere through mitosis resembled, and was partially dependent on, Nrc1 (Figures 3C & 4C). We therefore asked if the NCT (Nrc1 - CKII - TAFs) complex might mediate CKII-delivery, and thus whether the kinase is found in this context at specific genomic regions. To this end we used ChIP-seq to analyze three representative complex subunits (Nrc1, Cka1 and Taf7) and identified > 2,000 peaks for each factor (Figures 4D–E, S3–S5 & Table S2), with qPCR confirming the expected enrichment at ten test locations (Figures 4F & S4). Initial comparisons suggested a limited correlation between specific regions of Nrc1, Cka1 and Taf7 enrichment across the epigenetically complex centromere (Figures 4D & S3; see discussion), but a striking correspondence between all three factors along the chromosome arms (e.g. Figures 4E & S3). On considering the major peaks of Nrc1, Cka1 and Taf7 (~ top 15%: see Table S2), we noted that essentially all were located at transcription start sites (TSS ± 250 bp). Furthermore all three proteins were consistently co-enriched at these locations, with ~ 52, 54 and 47% of their respective major peaks in the context of NCT (Figure 5A). This strong overlap was maintained when all peaks associated with a TSS were considered, with the percentage of each within NCT > 42% in the case of Nrc1 and Cka1 (Figure 5B). Permutation testing (randomly sampling 106 TSS sets of equal size to those bound by Nrc1, Cka1 and Taf7) estimated the chance overlap rate for their major peaks at 1.43 ± 1.19 (208 observed; p < 10−6) and all peaks at 54.24 ± 6.96 (573 observed; p < 10−6). This highly significant disparity between chance and observation supported a direct relationship between all three factors.
Figure 5. NCT complex is enriched at the structural RNA genes.
(A–B) Nrc1, Cka1 and Taf7 are co-enriched at transcription start sites (TSS ± 250 bp). Peaks were identified, localized and quantified by optimized parameters (see Table S2). (C) 6,906* transcribed RNA species are annotated to the fission yeast chromosome arms (* excluding TEL3 and CEN1-3). These are classed by type and presented by percentage. (D–F) The structural RNAs are overrepresented as NCT peaks. Data is presented as the percentage of NCT peaks (major / all) per feature, and percentage coverage of each feature (specific NCT peaks were assigned to all overlapping features when required). For F the Fisher’s exact test (by R) was used to estimate the probability of the observed enrichment at structural RNAs: ‘Major NCT peaks’, p-value 5.5 × 10−16; ‘All NCT peaks’, p-value < 2.2 × 10−16 (the smallest value R will provide). (G) Nrc1, Cka1 and Taf7 enrichment across the histone genes (see also Figure S6). Each 10kb test window is named for the centered histone locus. In each case TBP and Taf7 are most enriched at the intergenic promoter region, while Nrc1 and Cka1 bind across the gene (note the unidirectional spread at unpaired hta2+).
We considered that NCT could represent TFIID, a component of the RNA polymerase II (RNApII) pre-initiation complex: Bdf1 (a potential homolog of Nrc1), CKII, and the TAF proteins all co-purify in this context from budding yeast (Auty et al., 2004; Matangkasombut et al., 2000). However multiple lines of evidence identify NCT as a distinct complex. For example NCT is associated with 194 of the 215 genes encoding structural RNAs (i.e. 5S rRNA, snoRNA, snRNA and tRNA: Figure 5C–F), many of which are transcribed by RNApIII (Roberts et al., 2003). Furthermore Nrc1, Cka1, and Taf7 in comparison to TBP (and by extension, TFIID) are not restricted to promoters, and their major peaks cover a much greater area (~ 1.2 kb NCT vs. ~ 450 bp TBP: Figure S5). Indeed the peak structure of specific subunits may indicate how NCT could associate with specific regions. Of Nrc1, Cka1 and Taf7, the last most closely resembled TBP (e.g. Figure 4E). This was particularly obvious at the divergently transcribed histone loci, where TBP and Taf7 were most highly enriched over the central promoter region, with Nrc1 and Cka1 also abundant across both gene bodies (Figures 5G & S6). In contrast Nrc1 and Cka1 spreading was unidirectional along the unpaired hta2+ (Figure 5G). Thus the TAF subunits (represented by Taf7) may mediate NCT recruitment to promoter-bound TBP, and the complex could then ‘spread’ over a wider area via the Nrc1 bromodomains binding H4ac (and/or other acetyl marks), perhaps also promoted by active transcription. In this manner Nrc1 would stabilize, rather than target, the association of NCT with chromatin, making best use of the low-affinity BD - Kac interaction (e.g. Figure 3F).
Condensin and the NCT complex bind similar genomic locations, but not at the same time
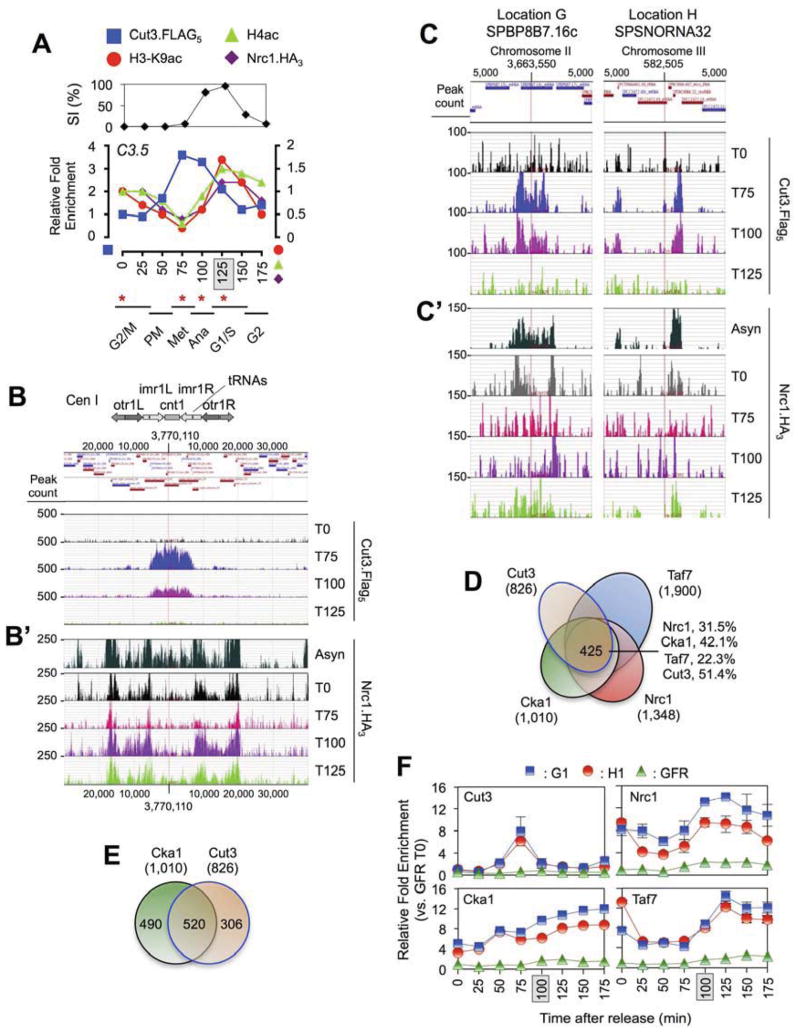
As above, mutants in NCT rescue those in condensin (e.g. Figures 2D, 3D & 4B). To further investigate if this indicated a direct regulatory relationship we tested if the two complexes shared a preference for locations additional to the core centromere (e.g. Figures 2D, 3C & 4C). Condensin is extra-chromosomal for much of the cell cycle (Nakazawa et al., 2008), so we used cell-cycle synchronized ChIP-seq of identify sites of complex binding (represented by the Cut3 subunit) at various stages through mitosis. As expected, Cut3 was highly enriched at the core centromere in metaphase / anaphase and then rapidly dissociated (Figures 6A–B & S3D). Cut3 associated with comparable kinetics at ~ 1,759 sites across the chromosome arms, including its previously reported preference for the structural RNAs (D’Ambrosio et al., 2008; Haeusler et al., 2008; Tanaka et al., 2012) (144 of the 215 genes; p-value < 2.2 × 10−16 : Figures 6C, S3H, S7 & Table S2). Of note the specific areas of Cut3 enrichment across the chromosome arms were strongly reminiscent of those bound by NCT (e.g. Figures S3 & S6). As an example Cut3, Nrc1, Cka1 and Taf7 share a preference for 425 TSS-containing regions (Figure 6D); > 60-fold more than expected by chance (permutation testing 106 events: 6.49 ± 2.52). Furthermore the overlap between [Cut3 : NCT] at these locations encompassed > 80% of [Cut3 : Cka1 alone] (Figure 6E), suggesting the central importance of NCT to any relationship between CKII and condensin.
Figure 6. Condensin and the NCT complex bind similar locations but are temporally anti-correlated through mitosis.
(A) Cells were synchronized (peak septation shaded), ChIPed (as in Figure 2C) and samples from the indicated time points (*) used to prepare ChIP-seq libraries. Cell-cycle stages are predictions: PM, prometaphase. (B, B′) Cut3 and Nrc1 binding across the chromosome 1 centromere are mitotically regulated (see also Figure S3): Cut3 levels are highest at metaphase (T75) when Nrc1 levels are at their lowest. ChIP-seq data from synchronized populations (time-points as in panel A) was loaded into Genplay as individual tracks relative to the annotated Sp genome. Asyn, Asynchronous. (C, C′) Cut3 and Nrc1 binding at similar locations across the chromosome arms are mitotically regulated (see also Figure S3). Note the different Y-axis scales in panels B and C. (D – E) The NCT (asynchronous; TSS ± 250 bp) and condensin (metaphase; TSS ± 500 bp) occupy similar locations. Cka1 is found in the context of NCT at > 80% of these [Cka1+ / condensin+] sites (425/520). (F) Cut3, Nrc1, Taf7 and Cka1 binding are mitotically regulated (determined by qPCR using the primers in Figure 4D–E). Samples were prepared from synchronized populations and all data (mean ± SD) normalized relative to the respective GFR-T0 (set to 1).
Finally we asked if condensin and NCT co-occupy their preferred locations along the chromosome arms through mitosis. To this end we subjected Nrc1 to synchronous ChIP-seq and compared its enrichment to that of Cut3 at each cell-cycle stage. As expected from direct ChIP analyses (e.g. Figures 2D & 6A), Nrc1 levels across the centromere were lowest at metaphase (T75) when Cut3 binding was at its peak (Figures 6B, B′ & S3). However a similar pattern was observed across the chromosome arms, with Nrc1 delocalizing from its preferred sites through mitosis and returning in G1/S (Figures 6C′ & S3). This profile extended through the NCT complex, with Nrc1, Cka1 and Taf7 occupancy at representative sites increasing as cells pass through mitosis (Figures 6F & S4). We note that such a pattern would co-localize CKII with its potential condensin substrate as cells complete mitosis and chromosome decondensation occurs (Figure 7).
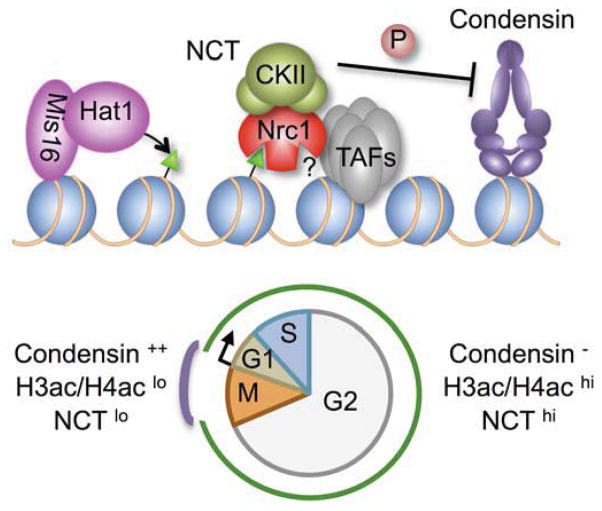
Figure 7. NCT as a condensin regulator.
H3ac/H4ac at the core centromere are catalyzed by the Hat1-Mis16 acetyltransferase. H4ac is recognized by Nrc1-BD1 to stabilize the association of NCT (Nrc1-CKII-TAFs) with chromatin. CKII phosphorylates condensin to inhibit its supercoiling activity (Takemoto et al., 2006). Though NCT and condensin share a preference for many chromosomal locations, their binding is regulated to anti-correlate through mitosis: this gives potential control over genome condensation and decondensation.
DISCUSSION
In this study we uncover and characterize a potential mechanism for the regulation of genome condensation at mitosis. We identify the Hat1-Mis16 acetyltransferase complex, show that this contributes to the global acetylation of histones H3 and H4, and establish its role in the mitotic regulation of these marks at the centromere (Figure 2). We demonstrate the selectivity of Nrc1-BD1 for H4ac and show that Nrc1 requires its bromodomains for efficient recruitment to the centromere (Figure 3). We then characterize the NCT (Nrc1-CKII-TAFs) complex (Figures 4 & 5), and present several lines of evidence that Hat1-Mis16 and NCT regulate the functionality of mitotic chromosomes via the condensin complex. This includes the ability of mutants in hat1, mis16, nrc1 and CKII to restore segregation-competent chromosomes when condensin is defective (e.g. Figures 1, 2G, 3D, 4B, S1A & S1K), and the extensive co-localization of NCT with condensin (e.g. Figures 6, S3 & S6). However the latter is not constitutive: NCT and condensin primarily overlap in early and late mitosis, and it is tempting to speculate that this temporally limited co-localization is directly regulatory to genome condensation at these cell-cycle stages (Figure 7).
The regulation and role(s) of NCT complex
Our identification of the NCT complex is supported by three independent approaches: co-purification, shared mutant phenotypes, and extensive subunit co-localization (e.g. Figures 4 & 5). Given its tandem bromodomains, it is likely that the Nrc1 subunit contributes to NCT targeting and / or binding at specific genomic locations (Filippakopoulos et al., 2012; Wang et al., 2013). In this regard, we show that both Nrc1 BDs are functional (Figure 3D) and required for efficient Nrc1 recruitment to the centromere (Figure 3C). Furthermore BD1 shows selective binding to the hyperacetylated H4 N-terminus (Figures 3E–F & S2E–G), and Nrc1 levels track centromeric H4ac levels through mitosis (Figure 2D). However the low affinity of this interaction (Figure 3F) suggests that additional, as yet unidentified, entities (e.g. linked to active transcription; Figure 5G) contribute to the efficient in vivo association of NCT with chromatin. These could be recognized by other members of the NCT complex at specific genomic locations and / or cell cycle stages.
This study does not specifically address any role(s) for NCT in interphase cells, although its specific sites of enrichment are highly suggestive of function. For example, the almost total coverage of structural RNAs (194 of the 215 genes: Figure 5C–F) indicates the importance of NCT to these locations, where the concomitant recruitment of CKII may contribute to the positive regulation of RNApIII transcription (Ghavidel and Schultz, 2001). It also remains to be determined if NCT functions exclusively as a CKII-delivery platform, although it is almost certainly not the only means to recruit this promiscuous kinase to chromatin. CKII is targeted to distinct subcellular locations by a range of associated proteins (Filhol and Cochet, 2009), and NCT likely differentially contributes to the total kinase pool at various chromosomal locations: this would help to explain the peak patterns of Cka1 versus Nrc1 and Taf7 at distinct regions, including the core centromere (Figures 4D & S3), and the only partial dependence of Cka1 on Nrc1 at this location (Figure 4C).
NCT as a regulator of condensin function
We have largely focused on the potential role of NCT in mitotic genome function, and considered a range of possible mechanisms by which the transient overlap between NCT and condensin in early and late mitosis could directly regulate chromosome condensation / decondensation. Recent studies show that human Brd4-isoform B mediates the recruitment of condensin II during DNA damage (Floyd et al., 2013). This is of particular note since Brd4 is the closest human homolog of Nrc1: both contain tandem bromodomains, share a specificity for H4ac, and are selectively inhibited by JQ1 ((Filippakopoulos et al., 2012; Filippakopoulos et al., 2010) and not shown). However Nrc1 does not contain the responsible C-terminal condensin II-interaction domain in Brd4-isoform B, Sp only contains the condensin I complex, and the suppressive genetic interactions of nrc1 and condensin mutants do not support a positive role for Nrc1 in condensin recruitment. In an alternate interpretation, their genetic interactions and mitotically-regulated binding profiles could indicate that NCT and condensin compete for occupancy at specific locations, such that NCT must be removed at prometaphase to allow condensin to load, and its return after anaphase displaces condensin. However evidence argues against such a relationship: NCT and condensin bind related rather than identical sites (e.g. compare the peaks in Figures S3 & S6), and Cut3 recruitment is comparable in WT and nrc1 cells (e.g. Figure 3C).
We rather favor a mechanism that accommodates all our findings and builds on the observation that human condensin I is phosphorylated, and thus catalytically inhibited, by CKII (Takemoto et al., 2006). In this manner any co-localization of kinase and substrate would be directly regulatory, such that the dissociation of NCT at early mitosis would relieve any CKII-mediated inhibition of condensin to control chromosome condensation, while its return in G1 would contribute to decondensation (Figure 7). This model would also accommodate the extensive literature relating mitotic kinase activity to condensin function (reviewed in (Bazile et al., 2010)).
Human condensin I is constitutively hyperphosphorylated, though the specific sites and responsible enzymes differ at interphase and mitosis. Phosphorylation at interphase (when condensin is primarily cytoplasmic) is mediated by CKII, while that at mitosis (after transport into the nucleus) is mediated by the mitotic kinases Cdk1 (Cdc2), Aurora B (Ipl1) and Polo (Cdc5) (Bazile et al., 2010; St-Pierre et al., 2009; Takemoto et al., 2006; Takemoto et al., 2007). The primary role of phosphorylation may be to control the catalytic activity of condensin, whose intrinsic ability to supercoil DNA is inhibited by CKII but activated by the mitotic kinases (Bazile et al., 2010). Of particular importance, the CKII-dependent inhibition of condensin is dominant over its Cdk1-dependent activation (Takemoto et al., 2006). This could explain how budding yeast condensin can remain constitutively associated with chromosomes (D’Ambrosio et al., 2008; Wang et al., 2005): their condensation at mitosis is not the direct consequence of condensin binding, but rather occurs after appropriate activation.
Chromosome condensation initiates in early mitosis, suggesting that condensin is fully activated in response to low CDK1 levels (Bazile et al., 2010). Spurious activation is a possible consequence of such ultrasensitivity, so condensin arrives from the cytoplasm painted with CKII-mediated inhibitory phosphorylations (Bazile et al., 2010; Takemoto et al., 2006). However we now show that CKII (both within and independent of the NCT) also occupies many condensin binding sites at the core centromere and chromosome arms (e.g. Figures 6C–E & S3). Any co-localization of inhibitory CKII with condensin at early mitosis would allow NCT to regulate the initiation of condensation, and could explain the increased rate of defective anaphases in nrc1Δ and ckb1Δ cells (Figures 1D & S1K).
As mitosis proceeds, full condensin activation / chromosome condensation would require NCT to dissociate from chromatin, and any inhibitory CKII-mediated phosphorylations to be removed (possibly by PP2A or PP1 phosphatases (Vagnarelli et al., 2006; Xing et al., 2008)). The precise means by which NCT is displaced prior to metaphase is unknown, although it may be a combination of mitotic phosphorylation (which drives the global displacement / relocalization of chromatin associated factors (Zaidi et al., 2010)) and reduced acetylation (Kruhlak et al., 2001; Sasaki et al., 2009) (Figures 2D & S1I). Of note, NCT does not entirely dissociate from chromatin, and indeed Cka1 levels on the chromosome arms gradually climb through mitosis from a nadir at G2/M (Figure 6F). However CKII is also phosphorylated, and inhibited, by CDK1 (Litchfield et al., 1992; St-Denis et al., 2009), so any co-localization of CKII with condensin through metaphase may have limited impact.
The means by which chromosome decondensation occurs at mitotic exit is poorly understood, although the kinetics of NCT return to condensin enrichment sites (Figure 6F) could suggest its role in this process. The return of NCT likely results from a combination of reduced mitotic kinase levels (Zaidi et al., 2010), increased histone acetylation (Kruhlak et al., 2001; Sasaki et al., 2009) (Figures 2D & S1), and the increased availability of Nrc1 (whose transcription peaks in G1 (Peng et al., 2006)). This would localize CKII with condensin after chromosome separation, allowing the kinase to inactivate the ATPase complex and directly promote decondensation. Studies to address each of these predictions are currently underway, but will require the development of approaches to monitor the condensation of specific locations at various mitotic stages.
EXPERIMENTAL PROCEDURES
Materials
Antibodies, yeast strain genotypes and ChIP oligonucleotides are in Table S3.
Bromodomain binding to histone peptide arrays
To produce recombinant forms of Nrc1, a region encompassing both bromodomains (residues S199-G526) was C-terminally fused to GST in pGEX-4T, heterologously expressed in E. coli, and purified by glutathione-sepharose affinity chromatography (Fuchs et al., 2010). Recombinant proteins (WT; BD1* (PDYP266–269AAAA); BD2* (PDYF428–431AAAA); or BD1*/BD2*) were used to probe a histone-tail peptide array with combinations of post-translational modifications and areas of enrichment identified from heat maps of the normalized mean intensity. All data was generated from a minimum of two arrays with 12 – 24 individual spots per peptide (Figures 3E & S2E–G). Array preparation, binding conditions and data analysis were as previously (Fuchs et al., 2010; Rothbart et al., 2012).
Cell-cycle synchronization
Cell-cycle synchronization, ChIP and septation analyses were performed after arrest by cdc25-22 (Kim et al., 2009) (Figure S1B).
ChIP-seq
Input or ChIP samples were converted to barcoded libraries by adaptor addition (Quail et al., 2008). In brief each sample was quantified, end-repaired (End-It kit, Epicenter Biotechnologies), an A-overhang added, and ligated to one of a set of 12 barcode-specific adaptor primers. The resulting libraries were size selected (600 ± 50 bp), amplified by 18 cycles of PCR, purified with SPRI beads (Agencourt AMPure XP; Fisher Scientific), quantified on a high-sensitivity DNA chip by 2100 bio-analyzer (Agilent Technologies, Inc.), mixed at equivalent concentrations, and multiplexed at 12 samples per Illumina HiSeq 2000 lane.
Data images from 100 bp runs (single or paired-end) were processed by the Illumina Sequence Control and Pipeline packages. ChIP-seq data was loaded into Genplay (v533) as individual tracks (50bp windows; normalized and INPUT subtracted) relative to the annotated fission yeast genome (Sanger Center release pombe090511) (Lajugie and Bouhassira, 2011; Zang et al., 2009). The online version of this paper links to a Genplay project to provide a dynamic visualization of the discussed tracks. Peaks for comparison and assignment to genomic features were automatically identified / quantified by optimized parameters (see Table S2). The location, level and timing of each peak, as well as their association with different features, was described by summary statistics and standard modeling approaches (using Microsoft Access and R). Permutation testing was used to measure the chance overlap rate between specific sets of ChIP-seq peaks by randomly sampling 106 sets of equal size to those under test.
In situ gene-tagging and mutagenesis
Constructs for epitope tagging were assembled by PCR megapriming from Sp genomic DNA or plasmid templates (Janke et al., 2004; Keogh et al., 2002), with products transformed / targeted by homologous recombination in the appropriate fission yeast backgrounds. Mutations at the Nrc1 bromodomains (BD1*, PDYP266–269AAAA; BD2*, PDYF428–431AAAA) were created by two-step marker replacement (Mehta et al., 2010). Nrc1 levels were modulated by ‘knocking-in’ alternate promoters of predicted expression outputs: fba1pr > lys4pr > ade4pr (Hiraoka et al., 2009) and the resulting expression determined by immunoblotting (Figure S2A).
Isothermal Titration Calorimetry
ITC measurements were conducted at 25°C with a MicroCal Auto-ITC200 in 1× PBS, pH 7.6 (2.7 mM KCl, 1.5 mM KH2PO4, 140 mM NaCl, 8 mM Na2HPO4). Recombinant forms of the Nrc1 bromodomains (residues S199-G526: see Figure 3A) were dialyzed in 1× PBS, pH 7.6 and brought to 30 μM. Peptides were lyophilized and resuspended in 1× PBS, pH 7.6 at 400 μM. Titrations were performed with 20 × 2 μl peptide injections at 180-second intervals using a reference power of 8 μcal/s. Results were analyzed using the MicroCal Origin software, and the Isotherm (μcal/sec) and integrated data (Kcal/Mole of injectant) for each titration presented (Figure 3F).
Microscopic analysis of mitotic chromosomes
Cells were grown in rich media and briefly (< 30 sec) fixed in 100% methanol at −80°C. DNA and microtubules were visualized by DAPI staining and anti-Tubulin immunoblotting. Anaphases were scored as defective if chromatin was observed near the metaphase plate after the bulk of separation had occurred (Kim et al., 2009). Data was presented as the percentage (mean ± s.d.) of late anaphase cells with chromosome segregation defects (counted > 100 cells per population: e.g. Figure 1D).
PEM-2 screening to identify second-site suppressors of condensin mutants
The PEM-2 approach (Roguev et al., 2008) was used to place ts alleles of each condensin ATPase subunit (cut3-477 [S1146P] or cut14-208 [S861P] (Sutani and Yanagida, 1997)) in the context of 3,002 fission yeast gene deletions (~ 75% of the non-essential Sp genome). Viable double-mutants were isolated at a permissive temperature for each condensin allele (25°C) and suppressors identified by replica-plating and improved growth at restrictive temperatures (34°C and 37°C: e.g. Figure 1B). To identify deletions that might suppress condensin by a non-specific mechanism parallel screening was performed with cdc25-22. Specific suppressors of interest were confirmed by direct crossing to a range of condensin alleles (cut3-477, cut14-208 and cnd2-1 [A114T]), tetrad dissection, and spot testing (e.g. Figure 1C) prior to further investigation.
Protein complex purification / identification of by LC-MS
C-terminally TAP-tagged proteins (yfg1+-CBP-TEV-2xProtein A) were purified and associated proteins identified by LC-MS as previously (Kim et al., 2009; Silva et al., 2012).
Protein immunoprecipitations
Cultures were grown to an OD600nm ~ 1.0 and pelleted for the preparation of whole cell extracts (WCEs). Cells were disrupted with glass beads in pre-chilled (4°C) lysis buffer (20mM Hepes pH 7.6, 200mM KOAc, 10% Glycerol, 1mM EDTA) supplemented with protease inhibitors (1mM PMSF, 1 μg/ml aprotinin, 1 μg/ml leupeptin and 1 μg/ml pepstatin-A). WCE samples were quantified, standardized for concentration, and nrc1.HA3 or Hat1.FLAG5 immunoprecipitated with anti-HA (12CA5) or anti-FLAG (FLAG-M2; Sigma) as previously (Keogh et al., 2002).
qPCR
ChIP samples were prepared as above and qPCR data obtained with an iCycler (Bio-Rad), SYBR green (Molecular Probes), Platinum Taq (Invitrogen) and the primer sets in Table S3. Quantitations were as previously (Silva et al., 2012), with all enrichments expressed as a percentage of input or relative to a gene-free region (GFR: Chromosome II 1,149,380 – 1,149,504) that shows no specific enrichment of TBP, Nrc1, Cka1, Taf7 or Cut3 in ChIP-seq (Figure S3).
Whole cell extracts for immunoblotting
WCEs were isolated with Trichloroacetic Acid (Mehta et al., 2010) and supernatants analyzed by immunoblotting.
Supplementary Material
HIGHLIGHTS: CELL-REPORTS-D-13-00300R3.
H3 / H4 acetylation at the centromere are Hat1-dependent and mitotically regulated
The Nrc1 bromodomain subunit of NCT shows a preference for hyperacetylated H4
NCT and condensin briefly co-localize during chromosome condensation and decondensation
Mutant Hat1 / NCT rescue mutant condensin and improve mitotic chromosome structure
Acknowledgments
We thank Steve Buratowski, Julien Lajugie, Andrew McLellan, Jamie Moseley, Charles Query, Mike Shales, Arthur Skoultchi, Jon Warner and Ian Willis for help and advice; Robin Allshire, Mark Bedford, James Bradner, Dan Finley, Keith Gull, Tetsuro Kokubo and Richard Maraia for the generous supply of materials. C.J.R. was supported by ICON plc and the UCD Newman Fellowship Program. J.F. was supported by start-up funding from Ryerson University and a discovery grant from the Natural Sciences and Engineering Research Council of Canada. T.K. is a Canada Research Chair in Proteomics of Cancer Research. This work was also supported in part by the National Institutes of Health (ES019966, GM085394, GM68088, GM85394); the Canadian Institutes of Health Research (MT-6092); NYSTEM, the funding agency of the Empire State Stem Cell Board (C024405 and C024172); pilot funding from the AECOM Epigenomics facility; and an NCI Cancer Center Support grant to Albert Einstein College of Medicine (CA013330).
Footnotes
ACCESSION NUMBERS
All sample data have been deposited in NCBI’s Gene Expression Omnibus (Edgar et al., 2002) and are accessible through GEO series accession number GSE53955.
Supplemental information including eight figures, three tables, and a Genplay project to visualize all ChIP-seq data can be found with this article online at http://dx.doi.org/XXXX.
AUTHOR CONTRIBUTIONS
R.M. and S.B.R. contributed equally to this work. H-S.K., R.M., S.B.R., V.V., T.K., N.J.K., J.S.F., B.D.S, W.E. and M-C.K. conceived the experiments. H-S.K., R.M., S.B.R., A.C.S., V.V., E.R., T.K., A.R., C.J.R., J.W., H.J. and J.S.F. performed the experiments. H-S.K., R.M., S.B.R, A.C.S., V.V., T.K., C.J.R., K.G.H, J.F.G, N.J.K, B.D.S., E.E.B., W.E. and M-C.K. analyzed the data. H-S.K., R.M., S.B.R. and M-C.K. wrote the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Auty R, Steen H, Myers LC, Persinger J, Bartholomew B, Gygi SP, Buratowski S. Purification of active TFIID from Saccharomyces cerevisiae. Extensive promoter contacts and co-activator function. J Biol Chem. 2004;279:49973–49981. doi: 10.1074/jbc.M409849200. [DOI] [PubMed] [Google Scholar]
- Bazile F, St-Pierre J, D’Amours D. Three-step model for condensin activation during mitotic chromosome condensation. Cell Cycle. 2010;9:3243–3255. doi: 10.4161/cc.9.16.12620. [DOI] [PubMed] [Google Scholar]
- Beltrao P, Trinidad JC, Fiedler D, Roguev A, Lim WA, Shokat KM, Burlingame AL, Krogan NJ. Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species. PLoS Biol. 2009;7:e1000134. doi: 10.1371/journal.pbio.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson LJ, Phillips JA, Gu Y, Parthun MR, Hoffman CS, Annunziato AT. Properties of the type B histone acetyltransferase Hat1: H4 tail interaction, site preference, and involvement in DNA repair. J Biol Chem. 2007;282:836–842. doi: 10.1074/jbc.M607464200. [DOI] [PubMed] [Google Scholar]
- Chen ES, Sutani T, Yanagida M. Cti1/C1D interacts with condensin SMC hinge and supports the DNA repair function of condensin. Proc Natl Acad Sci USA. 2004;101:8078–8083. doi: 10.1073/pnas.0307976101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cuylen S, Haering CH. Deciphering condensin action during chromosome segregation. Trends Cell Biol. 2011;21:552–559. doi: 10.1016/j.tcb.2011.06.003. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C, Schmidt CK, Katou Y, Kelly G, Itoh T, Shirahige K, Uhlmann F. Identification of cis-acting sites for condensin loading onto budding yeast chromosomes. Genes Dev. 2008;22:2215–2227. doi: 10.1101/gad.1675708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhalluin C, Carlson JE, Zeng L, He C, Aggarwal AK, Zhou MM. Structure and ligand of a histone acetyltransferase bromodomain. Nature. 1999;399:491–496. doi: 10.1038/20974. [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filhol O, Cochet C. Protein kinase CK2 in health and disease: Cellular functions of protein kinase CK2: a dynamic affair. Cell Mol Life Sci. 2009;66:1830–1839. doi: 10.1007/s00018-009-9151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Picaud S, Mangos M, Keates T, Lambert JP, Barsyte-Lovejoy D, Felletar I, Volkmer R, Müller S, Pawson T, et al. Histone recognition and large-scale structural analysis of the human bromodomain family. Cell. 2012;149:214–231. doi: 10.1016/j.cell.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floyd SR, Pacold ME, Huang Q, Clarke SM, Lam FC, Cannell IG, Bryson BD, Rameseder J, Lee MJ, Blake EJ, et al. The bromodomain protein Brd4 insulates chromatin from DNA damage signalling. Nature. 2013;498:246–250. doi: 10.1038/nature12147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs SM, Krajewski K, Baker RW, Miller VL, Strahl BD. Influence of combinatorial histone modifications on antibody and effector protein recognition. Curr Biol. 2010;21:53–58. doi: 10.1016/j.cub.2010.11.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghavidel A, Schultz MC. TATA binding protein-associated CK2 transduces DNA damage signals to the RNA polymerase III transcriptional machinery. Cell. 2001;106:575–584. doi: 10.1016/s0092-8674(01)00473-1. [DOI] [PubMed] [Google Scholar]
- Haeusler RA, Pratt-Hyatt M, Good PD, Gipson TA, Engelke DR. Clustering of yeast tRNA genes is mediated by specific association of condensin with tRNA gene transcription complexes. Genes Dev. 2008;22:2204–2214. doi: 10.1101/gad.1675908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Fujita Y, Iwasaki O, Adachi Y, Takahashi K, Yanagida M. Mis16 and Mis18 are required for CENP-A loading and histone deacetylation at centromeres. Cell. 2004;118:715–729. doi: 10.1016/j.cell.2004.09.002. [DOI] [PubMed] [Google Scholar]
- Heale JT, Ball AR, Jr, Schmeising JA, Kim JS, Dong X, Zhou S, Hudson DF, Earnshaw WC, Yokomori K. Condensin I interacts with the PARP-1-XRCC1 complex and functions in DNA single-strand break repair. Mol Cell. 2006;21:837–848. doi: 10.1016/j.molcel.2006.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. Condensins: universal organizers of chromosomes with diverse functions. Genes Dev. 2012;26:1659– 1678. doi: 10.1101/gad.194746.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Kawamata K, Haraguchi T, Chikashige Y. Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009;14:499–509. doi: 10.1111/j.1365-2443.2009.01284.x. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes beta-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Hirota T, Gerlich D, Koch B, Ellenberg J, Peters JM. Distinct functions of condensin I and II in mitotic chromosome assembly. J Cell Sci. 2004;117:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- Jacobson RH, Ladurner AG, King DS, Tjian R. Structure and function of a human TAFII250 double bromodomain module. Science. 2000;288:1422–1425. doi: 10.1126/science.288.5470.1422. [DOI] [PubMed] [Google Scholar]
- Janke C, Magiera MM, Rathfelder N, Taxis C, Reber S, Maekawa H, Moreno-Borchart A, Doenges G, Schwob E, Schiebel E, Knop M. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter-substitution cassettes. Yeast. 2004;21:947–962. doi: 10.1002/yea.1142. [DOI] [PubMed] [Google Scholar]
- Johzuka K, Horiuchi T. The cis element and factors required for condensin recruitment to chromosomes. Mol Cell. 2009;34:26–35. doi: 10.1016/j.molcel.2009.02.021. [DOI] [PubMed] [Google Scholar]
- Keogh MC, Cho EJ, Podolny V, Buratowski S. Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol Cell Biol. 2002;22:1288–1297. doi: 10.1128/mcb.22.5.1288-1297.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–623. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Vanoosthuyse V, Fillingham J, Roguev A, Watt S, Kislinger T, Treyer A, Carpenter LR, Bennett CS, Emili A, et al. An acetylated form of histone H2A.Z regulates chromosome architecture in Schizosaccharomyces pombe. Nat Struct Mol Biol. 2009;16:1286–1293. doi: 10.1038/nsmb.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura K, Rybenkov VV, Crisona NJ, Hirano T, Cozzarelli NR. 13S condensin actively reconfigures DNA by introducing global positive writhe: implications for chromosome condensation. Cell. 1999;98:239–248. doi: 10.1016/s0092-8674(00)81018-1. [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Hendzel MJ, Fischle W, Bertos NR, Hameed S, Yang XJ, Verdin E, Bazett-Jones DP. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- Lajugie J, Bouhassira EE. GenPlay, a multipurpose genome analyzer and browser. Bioinformatics. 2011;27:1889–1893. doi: 10.1093/bioinformatics/btr309. [DOI] [PubMed] [Google Scholar]
- Litchfield DW, Lüscher B, Lozeman FJ, Eisenman RN, Krebs EG. Phosphorylation of casein kinase II by p34cdc2 in vitro and at mitosis. J Biol Chem. 1992;267:13943–13951. [PubMed] [Google Scholar]
- Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK, et al. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508–512. doi: 10.1038/nature09272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matangkasombut O, Buratowski RM, Swilling NW, Buratowski S. Bromodomain factor 1 corresponds to a missing piece of yeast TFIID. Genes Dev. 2000;14:951–962. [PMC free article] [PubMed] [Google Scholar]
- Mehta M, Braberg H, Wang S, Lozsa A, Shales M, Solache A, Krogan NJ, Keogh MC. Individual lysine acetylations on the N-terminus of S. cerevisiae H2A.Z are highly but not differentially acetylated. J Biol Chem. 2010;285:39855–39865. doi: 10.1074/jbc.M110.185967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa N, Nakamura TM, Kokubu A, Ebe M, Nagao K, Yanagida M. Dissection of the essential steps for condensin accumulation at kinetochores and rDNAs during fission yeast mitosis. J Cell Biol. 2008;180:1115–1131. doi: 10.1083/jcb.200708170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onn I, Aono N, Hirano M, Hirano T. Reconstitution and subunit geometry of human condensin complexes. EMBO J. 2007;26:1024–1034. doi: 10.1038/sj.emboj.7601562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Fang Y, Spector DL, Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol Biol Cell. 2004;15:3296–3308. doi: 10.1091/mbc.E04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono T, Losada A, Hirano M, Myers MP, Neuwald AF, Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/s0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Ornaghi P, Yang JC, Lowe N, Evans PR, Ballario P, Neuhaus D, Filetici P, Travers AA. The structural basis for the recognition of acetylated histone H4 by the bromodomain of histone acetyltransferase Gcn5p. EMBO J. 2000;19:6141–6149. doi: 10.1093/emboj/19.22.6141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Karuturi RK, Miller LD, Lin K, Jia Y, Kondu P, Wang L, Wong LS, Liu ET, Balasubramanian MK, Liu J. Identification of cell cycle-regulated genes in fission yeast. Mol Biol Cell. 2006;16:1026–1042. doi: 10.1091/mbc.E04-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pidoux AL, Choi ES, Abbott JK, Liu X, Kagansky A, Castillo AG, Hamilton GL, Richardson W, Rappsilber J, He X, Allshire RC. Fission yeast Scm3: A CENP-A receptor required for integrity of subkinetochore chromatin. Mol Cell. 2009;33:299–311. doi: 10.1016/j.molcel.2009.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail MA, Kozarewa I, Smith F, Scally A, Stephens PJ, Durbin R, Swerdlow H, Turner DJ. A large genome center’s improvements to the Illumina sequencing system. Nat Methods. 2008;5:1005–1010. doi: 10.1038/nmeth.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DN, Stewart AJ, Huff JT, Cairns BR. The RNA polymerase III transcriptome revealed by genome-wide localization and activity-occupancy relationships. Proc Natl Acad Sci USA. 2003;100:14695–14700. doi: 10.1073/pnas.2435566100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. Conservation and rewiring of functional modules revealed by an epistasis map (E-MAP) in fission yeast. Science. 2008;322:405–410. doi: 10.1126/science.1162609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothbart SB, Lin S, Britton LM, Krajewski K, Keogh MC, Garcia BA, Strahl BD. Poly-acetylated chromatin signatures are preferred epitopes for site-specific histone H4 acetyl antibodies. Sci Rep. 2012;2:489. doi: 10.1038/srep00489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussou I, Draetta G. The Schizosaccharomyces pombe casein kinase II alpha and beta subunits: evolutionary conservation and positive role of the beta subunit. Mol Cell Biol. 1994;14:576–586. doi: 10.1128/mcb.14.1.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AL, Wang W, Graybosch DM, Li H, Allis CD, Patel DJ, Verdine GL. Histone H3 recognition and presentation by the WDR5 module of the MLL1 complex. Nat Struct Mol Biol. 2006;13:704–712. doi: 10.1038/nsmb1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CJ, Roguev A, Patrick K, Xu J, Jahari H, Tong Z, Beltrao P, Shales M, Qu H, Collins SR, et al. Hierarchical modularity and the evolution of genetic interactomes across species. Mol Cell. 2012;46:691–704. doi: 10.1016/j.molcel.2012.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarge KD, Park-Sarge OK. Mitotic bookmarking of formerly active genes: Keeping epigenetic marks from fading. Cell Cycle. 2009;8:818–823. doi: 10.4161/cc.8.6.7849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Ito T, Nishino N, Khochbin S, Yoshida M. Real-time imaging of histone H4 hyperacetylation in living cells. Proc Natl Acad Sci USA. 2009;106:16257–16262. doi: 10.1073/pnas.0902150106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva AC, Xu X, Kim HS, Fillingham J, Kislinger T, Mennella TA, Keogh MC. The Replication-Independent Chaperones HIR, Asf1 and Rtt106 co-operate to maintain promoter fidelity. J Biol Chem. 2012;287:1709–1718. doi: 10.1074/jbc.M111.316489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci USA. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis NA, Derksen RD, Litchfield DW. Evidence for regulation of mitotic progression through temporal phosphorylation and dephosphorylation of CK2alpha. Mol Cell Biol. 2009;29:2068–2081. doi: 10.1128/MCB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre J, Douziech M, Bazile F, Mirela Pascariu M, Bonneil E, Sauvé V, Ratsima H, D’Amours D. Polo kinase regulates mitotic chromosome condensation by hyperactivation of condensin DNA supercoiling activity. Mol Cell. 2009;34:416–426. doi: 10.1016/j.molcel.2009.04.013. [DOI] [PubMed] [Google Scholar]
- Steen RL, Cubizolles F, Le Guellec K, Collas P. A kinase-anchoring protein (AKAP)95 recruits human chromosome-associated protein (hCAP)-D2/Eg7 for chromosome condensation in mitotic extract. J Cell Biol. 2000;149:531–536. doi: 10.1083/jcb.149.3.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutani T, Yanagida M. DNA renaturation activity of the SMC complex implicated in chromosome condensation. Nature. 1997;388:798–801. doi: 10.1038/42062. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tada K, Susumu H, Sakuno T, Watanabe Y. Condensin association with histone H2A shapes mitotic chromosomes. Nature. 2011;474:477–483. doi: 10.1038/nature10179. [DOI] [PubMed] [Google Scholar]
- Takemoto A, Kimura K, Yanagisawa J, Yokoyama S, Hanaoka F. Negative regulation of condensin I by CK2-mediated phosphorylation. EMBO J. 2006;25:5339–5348. doi: 10.1038/sj.emboj.7601394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto A, Maeshima K, Ikehara T, Yamaguchi K, Murayama A, Imamura S, Imamoto N, Yokoyama S, Hirano T, Watanabe Y, et al. The chromosomal association of condensin II is regulated by a noncatalytic function of PP2A. Nat Struct Mol Biol. 2009;16:1302–1308. doi: 10.1038/nsmb.1708. [DOI] [PubMed] [Google Scholar]
- Takemoto A, Murayama A, Katano M, Urano T, Furukawa K, Yokoyama S, Yanagisawa J, Hanaoka F, Kimura K. Analysis of the role of Aurora B on the chromosomal targeting of condensin I. Nucl Acids Res. 2007;35:2403–2412. doi: 10.1093/nar/gkm157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Tanizawa H, Sriswasdi S, Iwasaki O, Chatterjee AG, Speicher DW, Levin HL, Noguchi E, Noma K. Epigenetic regulation of condensin-mediated genome organization during the cell cycle and upon DNA damage through histone H3 lysine 56 acetylation. Mol Cell. 2012;48:532–546. doi: 10.1016/j.molcel.2012.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trievel RC, Shilatifard A. WDR5, a complexed protein. Nat Struct Mol Biol. 2009;16:678–680. doi: 10.1038/nsmb0709-678. [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Hudson DF, Ribeiro SA, Trinkle-Mulcahy L, Spence JM, Lai F, Farr CJ, Lamond AI, Earnshaw WC. Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol. 2006;8:1133–1142. doi: 10.1038/ncb1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang BD, Eyre D, Basrai M, Lichten M, Strunnikov A. Condensin binding at distinct and specific chromosomal sites in the Saccharomyces cerevisiae genome. Mol Cell Biol. 2005;25:7216–7225. doi: 10.1128/MCB.25.16.7216-7225.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Tadeo X, Hou H, Tu PG, Thompson J, Yates JRr, Jia S. Epe1 recruits BET family bromodomain protein Bdf2 to establish heterochromatin boundaries. Genes Dev. 2013;27:1886–1902. doi: 10.1101/gad.221010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JS, Hayashi T, Yanagida M, Russell P. Fission yeast Scm3 mediates stable assembly of Cnp1/CENP-A into centromeric heterochromatin. Mol Cell. 2009;33:287–298. doi: 10.1016/j.molcel.2009.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AJ, Severson AF, Meyer BJ. Condensin and cohesin complexity: the expanding repertoire of functions. Nature Reviews Genetics. 2010;11:391–404. doi: 10.1038/nrg2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Vanderford NL, Sarge KD. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a Tool to Predict Kinase-specific Phosphorylation Sites in Hierarchy. Mol Cell Proteomics. 2008;7:1598–1608. doi: 10.1074/mcp.M700574-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi SK, Young DW, Montecino MA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Mitotic bookmarking of genes: a novel dimension to epigenetic control. Nat Rev Genet. 2010;11:583–589. doi: 10.1038/nrg2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W. A clustering approach for identification of enriched domains from histone modification ChIP-seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.