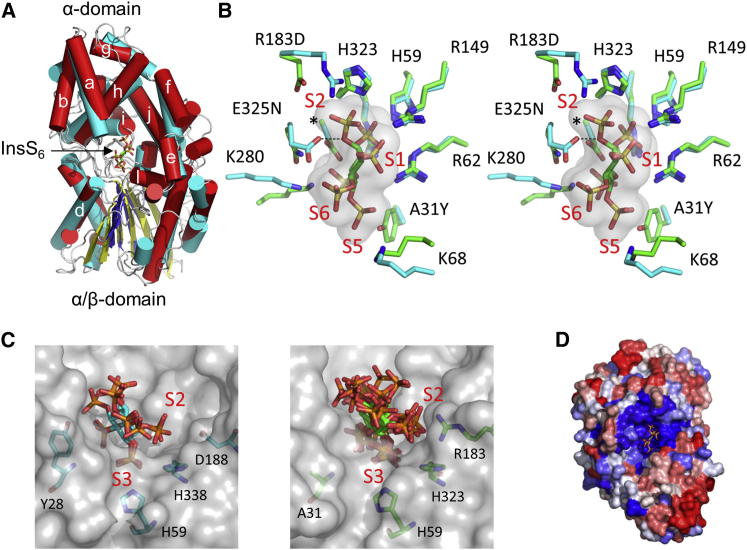
Figure 3.
X-Ray Crystal Structure of the BtMinpp
(A) Cylinder and ribbon diagram showing a superposition of the crystal structures of BtMinpp (α helices in red, β sheet in yellow) and the Aspergillus niger phytase PhyA (Oakley, 2010) (α helices in cyan, β sheet in blue) both taken from their complexes with the InsP6 analog, InsS6. The core α+β and capping α-domains are indicated. The naming of α helices in the capping domain of BtMinpp (indicated by a lowercase letter) follows that suggested for PhyA (Oakley, 2010). The atoms of the bound substrate analog are shown as sticks.
(B) A stereoview of the superposition of the side chains of InsS6-binding residues (shown as sticks) of BtMinpp (carbon atoms colored cyan) and Aspergillus niger PhyA (carbon atoms colored green). The residues selected for display form the primary sites of interaction with InsS6 in both complexes. The van der Waals surface of InsS6 is shown in gray. Residue labels follow the numbering for the BtMinpp structure. The leading character identifies the residue in BtMinpp, whereas the trailing identifies the corresponding variant residue found in PhyA. An asterisk (∗) indicates the position of the A324D substitution. Selected binding pockets (S1, S2, S5, and S6) are indicated, numbered according to corresponding sulfate group on the bound ligand. Pocket S4 is obscured in this orientation by the 6-sulfate group of the ligand. The interaction distance between a carboxyl group oxygen of residue E325 and the bridging ether oxygen of the S3 sulfate group of InsS6 is 3.3 Å and is indicated by a dashed line.
(C) Predicted binding modes within 1 kcal mol-1 of the minimum binding energy resulting from molecular docking experiments for InsP6 with PhyA (left, three modes) and BtMinpp (right, seven modes). Selected active site residues are shown as sticks and labeled as are binding pockets S2 and S3. The enzyme molecular surfaces are shown in gray.
(D) View of a ConSurf (Landau et al., 2005) color-coded surface representation of BtMinpp. The normalized conservation scores calculated by ConSurf are a relative measure of evolutionary conservation at each residue position based on the alignment of 23 MINPP sequences. The highest scores (8 and 9 on the ConSurf scale) represent the most conserved residue positions and are shown colored blue. The residues with the lowest scores (i.e., the most variable) are colored red. The bound InsS6 ligand is shown in stick format.