Fig. 2.
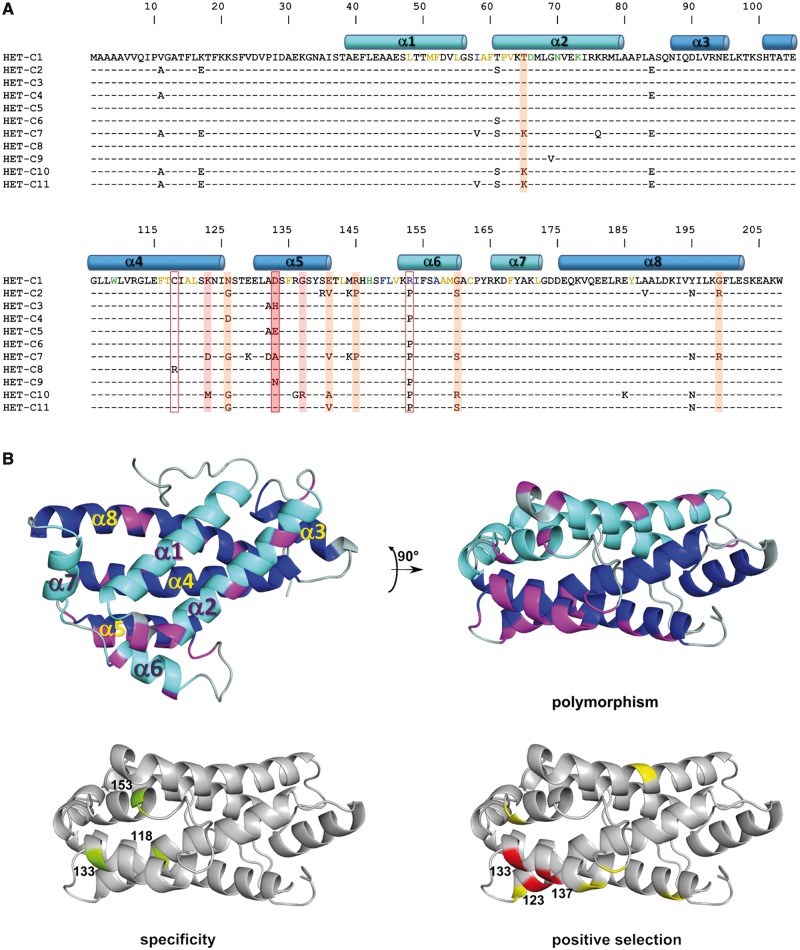
(A) Alignment of amino acid sequences encoded by the 11 known naturally occurring het-c alleles found in this study and previous studies. The amino acid sequence encoded by het-c1 is shown completely, only sequence differences are given for the other het-c alleles. Position of the α-helices of HET-C is given in cyan and blue, respectively, for the helices forming the two layers of the sandwich motif. Residues colored in yellow correspond to residues involved in lipid binding, in green to residues involved in sugar binding, and in blue to residues proposed to interact with membranes. Residues boxed in red (118, 133, 153) are shown to be involved in allele specificity. Residues shadowed in red and orange correspond to residues under positive selection with a 99% and 95% confidence level, respectively. (B) Three-dimensional structure of HET-C protein (Kenoth et al. 2010). The α-helices that form the two layers of the sandwich motif (α1, α2, α6, α7 and α3, α4, α5, α8) are given in cyan and blue, respectively. On the two top panels, polymorphic positions are given in magenta. The two lower panels identify the residues shown to be involved in allele specificity in green and the residues under positive selection in red for residues with a 99% confidence level and in yellow for residues with a 95% confidence level.