Fig. 2.
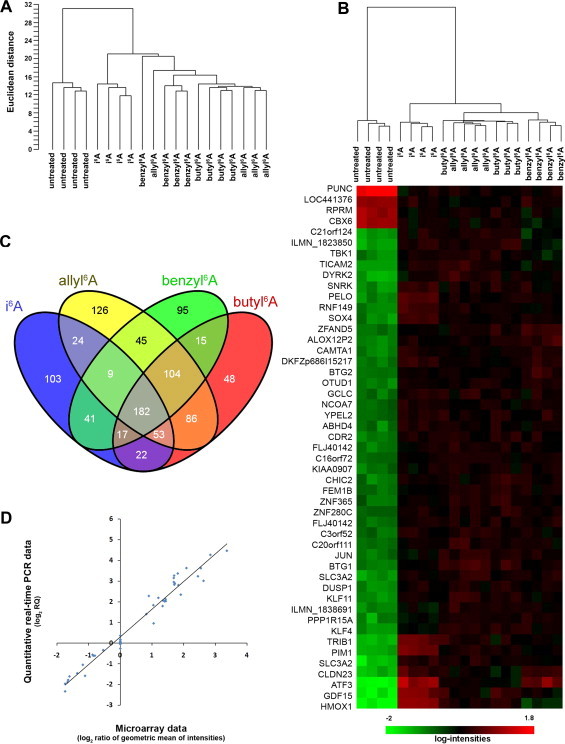
Gene expression profiles of untreated MCF7 cells and of cells treated for 6 h with 10 μM i6A or with equi-effective concentrations of allyl6A, benzyl6A or butyl6A. (A) Unsupervised clustering of samples (four replicates each) based on the expression levels of 3286 genes (detection P-value < 0.05 and coefficient of variation >0.15) revealed two main branches separating untreated from treated samples. Among treated cells, those treated with i6A clustered in a single branch distinct from those treated with the other three compounds. (B) Heatmap, resulted from the class comparison analysis, showing the first 49 most significantly (P < 1.0 × 10−10) differentially expressed genes in treated versus untreated cells and the clustering of samples (on top of the heatmap) based on the expression of these 49 genes only. Gene expression levels are indicated by the color bar: green, low; red, high. (C) Venn diagram of the numbers of differentially expressed genes (P < 1.0 × 10−4 and ≥1.5-fold) in MCF7 cells treated with i6A or one of its analogs, each compared to untreated cells. Overall, 182 genes were modified by all four nucleosides. (D) Correlation between microarray and quantitative PCR data for 9 genes measured under all five treatment conditions. Pearson’s r = 0.98, P < 0.0001.
