Figure 3. Metaproteomic comparison of human proteins in modern and ancient dental samples.
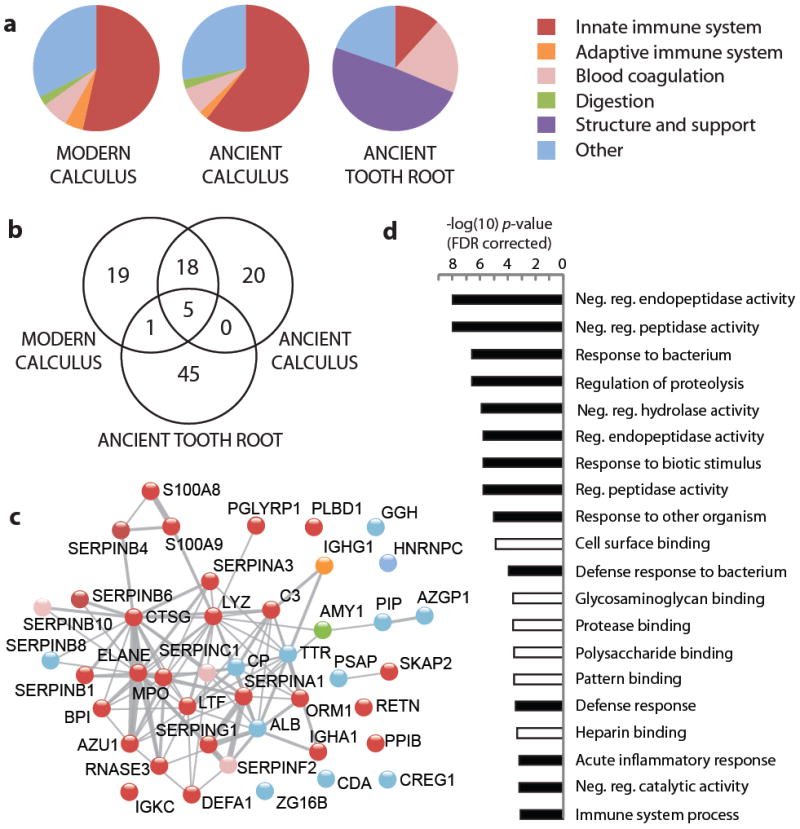
a, Functional characterization of human proteins identified in modern dental calculus (two individuals), ancient dental calculus (four individuals), and ancient tooth roots (four individuals). b, Venn diagram of shared human proteins by sample type. c, STRING network representation of human proteins identified in ancient dental calculus; nodes are labeled by gene name and colored in accordance with functional categories and connections (gray) to predicted functional partners. The network is set to medium confidence (0.4) for all active prediction methods. d, Gene ontology categories with significant enrichment (p < 0.001, FDR corrected) in ancient dental calculus for biological process (black) and molecular function (white) relate to proteinase regulation, substrate binding, and innate immune function. Enrichment calculated relative to the human genome using the STRING-embedded AmiGO term enrichment tool.
