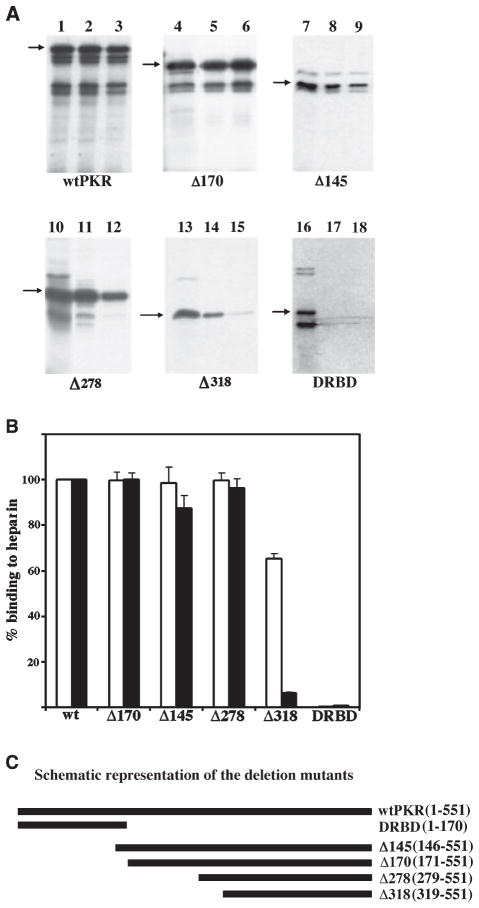
Fig. 1.
(A) Residues between 279 and 318 are important for heparin-binding activity of PKR. The wild-type PKR (wtPKR) and its deletion mutants were tested for heparin–agarose binding activity. In vitro translated proteins (5 μL) were bound to heparin–agarose in binding buffer and the proteins remaining bound to the beads after washing were analyzed by SDS/PAGE followed by phosphorimager analysis. Lanes 1, 4, 7, 10, 13 and 16 represent total proteins present in the translation mix. Lanes 2, 5, 8, 11, 14, and 17 represent proteins bound at 50 mM salt and lanes 3, 6, 9, 12, 15 and 18 represent proteins bound at 200 mM salt. The different proteins that were tested are as indicated at the bottom of the panels and positions of the proteins are indicated by arrows. Additional bands observed below the expected bands arise due to initiation of translation at internal AUG codons in rabbit reticulocyte system. (B) Quantification of heparin-binding activity of deletion mutants. The percentage binding of various deletion mutants was quantified by phosphorimager analysis. The binding activity of the wtPKR was taken as 100% and binding of mutants is represented relative to this value. The white bars represent binding at 50 mM salt concentration and the black bars represent the binding activity at 200 mM salt. Error bars represent SD calculated based on three experiments. (C) A schematic representation of the deletion mutants. The names of the mutants and the residues retained in each mutant are indicated on the right.