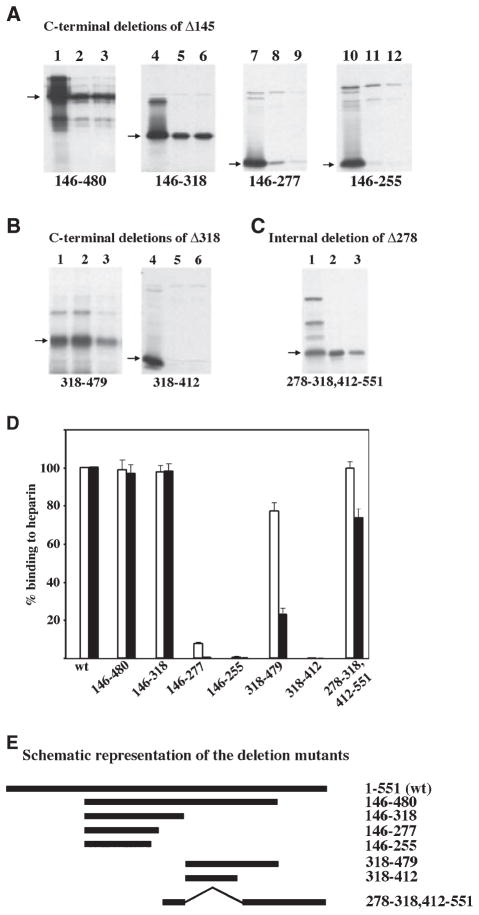
Fig. 2.
(A) Heparin-binding activity of carboxy terminal deletion mutants of Δ145. The heparin–agarose binding activity of carboxy terminal deletion mutants of Δ145 was tested as described in Fig. 1 legend. Lane 1, 4, 7 and 10 represent total proteins in the translation mix. Lanes 2, 5, 8 and 11, binding performed at 50 mM salt; lanes 3, 6, 9 and 12, binding performed at 200 mM salt. Arrows indicate the positions of deletion mutants and the labels at the bottom of the panels show the residues retained in the deletion mutants. (B) A second region between residues 413 and 479 also contributes to heparin-binding activity of PKR. The heparin–agarose binding activity of carboxy terminal deletion mutants of Δ318 was tested. Lane 1 and 4, total proteins in the translation mix; lanes 2 and 5, binding performed at 50 mM salt; lanes 3 and 6, binding performed at 200 mM salt. Arrows indicate the positions of deletion mutants and the labels at the bottom of the panels show the residues retained in the deletion mutants. (C) The region between 318 and 412 is dispensable for heparin-binding activity of PKR. An internal deletion mutant of Δ278 was created that lacked amino acids between 318 and 412 (278-ID). The heparin–agarose binding activity of this mutant was tested. Lane 1, total protein in the translation mix; lane 2, protein bound to heparin–agarose at 50 mM salt; lane 3, protein bound to heparin–agarose at 200 mM salt. An arrow indicates the position of the deletion mutant. (D) Quantification of heparin-binding activity of deletion mutants. The percentage binding of various deletion mutants was quantified by phosphorimager analysis. The binding activity of the wt PKR was taken as 100% and binding of mutants is represented relative to this value. The white bars represent binding at 50 mM salt concentration and the black bars represent the binding activity at 200 mM salt. Error bars represent SD calculated based on three experiments. (E) A schematic representation of the deletion mutants. The names of the mutants (residues retained in each mutant) are indicated on the right.