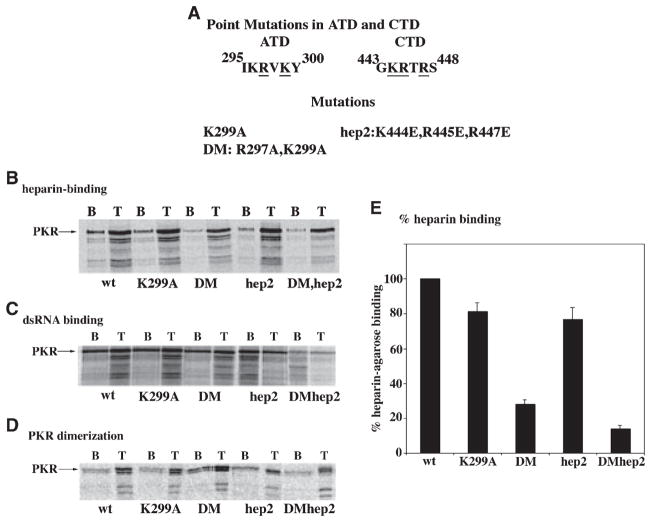
Fig. 5.
(A) Point mutations in ATD and CTD. The amino acids targeted by mutations within ATD and CTD are underlined. The individual mutations are as described in the text. (B) Heparin-binding activity of the mutants. The heparin–agarose binding activity of the point mutants was tested. The T lanes represent total proteins in the translation mix. The B lanes represent binding performed at 200 mM salt. Arrows indicate the positions of PKR mutants and the mutant name is shown at the bottom of the panels. (C) dsRNA-binding activity of the mutants. The dsRNA-binding activity of the point mutants was tested by their binding to poly(I)·poly(C)-agarose. The T lanes represent total proteins in the translation mix. The B lanes represent binding performed at 300 mM salt. Arrows indicate the positions of PKR mutants and the mutant name is shown at the bottom of the panels. (C) Dimerization activity of the point mutants. The ability of point mutants to dimerize was tested using the in vitro dimerization assay. The T lanes represent total proteins in the translation mix. The B lanes represent binding to PKR immobilized on Ni-charged His-bind resin performed at 200 mM salt. Arrows indicate the positions of PKR mutants and the mutant name is shown at the bottom of the panels. (D) Quantification of the heparin-binding activity of point mutants. The data shown in panel A was quantified using a phosphorimager analysis. The binding activities of mutants are represented as a percentage of wild-type PKR. The error bars represent SD from three separate experiments.