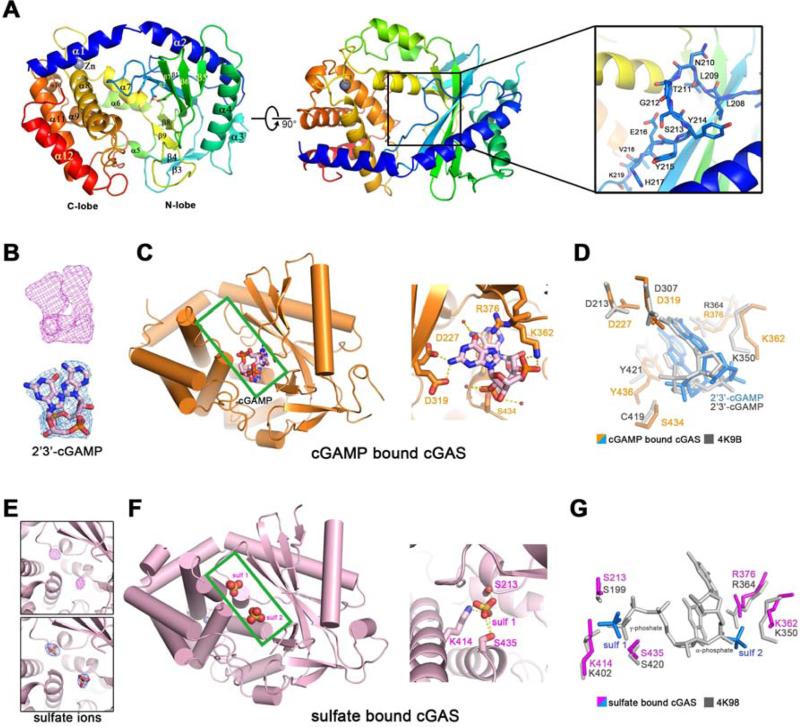
Figure 1. Overall structures of human cGAS in apo-form or in complex with 2’3’-cGAMP or sulfate ions.
(A) Overall structure of human cGAS in the apo-form. The catalytic residues are shown in sticks. Two perpendicular views and a close-up view of the conserved loop connecting the first and second β strand are shown. The residues in the loop are shown in sticks. (B) The simulated annealing omit map (upper panel) and the 2Fo-Fc electron density map (lower pannel) for 2’3’-cGAMP are contoured at 3.0 σ and 1.0 σ, respectively. (C) Overall structure of cGAS in complex with 2’3’-cGAMP (left panel). The cleft between N-lobe and C-lobe is highlighted by green rectangle. 2’3’-cGAMP binds to cGAS by multiple polar contacts (right panel). (D) Residues involved in 2’3’-cGAMP binding are identical to the ones in mouse-cGAS-DNA-2’3’-cGAMP ternary complex (PDB: 4K9B). (E) The simulated annealing omit map (upper panel) and the 2Fo-Fc electron density map (lower panel) for the sulfate ion are contoured at 4.0 σ and 1.5 σ, respectively. (F) Overall structure of cGAS in complex with sulfate ions (left panel). The cleft between N-lobe and C-lobe is highlighted by green rectangle. Sulfate ion 1 is coordinated by several polar residues (right panel). (G) Residues involved in sulfate ion binding are conserved in mouse-cGAS-DNA-linear-2’-GTP-GMP ternary complex (PDB: 4K98). The position of sulfate ion 1 is similar to the γ-phosphate group of GTP moiety, whereas the position of sulfate ion 2 is similar to the α-phosphate group of GMP moiety. All the structure figures are prepared in PyMol (DeLano, 2002).