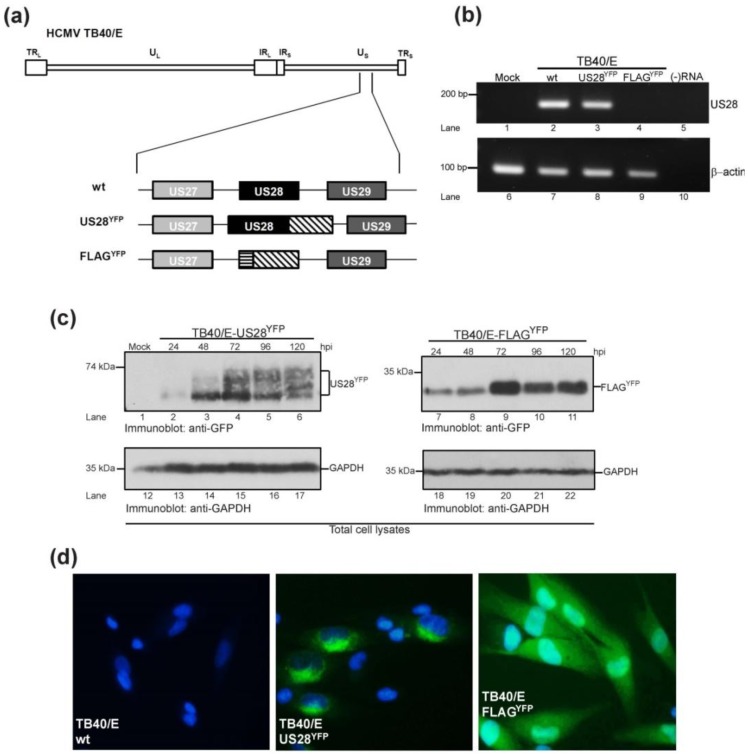
Figure 1.
Generation of TB40/E-US28 variants. (a) Using a bacterial artificial chromosome (BAC) recombineering approach Human cytomegalovirus (HCMV) TB40/E variants were generated that express either chimeric US28 containing a carboxy-terminal YFP tag (US28YFP) or a US28 deletion mutant where the US28 ORF has been replaced with an engineered FLAG-YFP cassette (FLAGYFP). YFP sequences are denoted by the diagonally hatched box; FLAG sequences are denoted by the horizontally striped box. TR, terminal repeat; U, unique sequences; IR, inverted repeat; L, long; S, short. (b) Fibroblasts mock-infected or infected (MOI = 5) with TB40/E wt or TB40/E-US28 variants were harvested 48 hours post-infection and subjected to RT-PCR with primers specific to US28 (lanes 1–5) or β-actin (lanes 6–10). A sample lacking RNA ((−)RNA) was included as a negative control. HCMV US28, β-actin, and relative DNA standards are indicated. (c) Fibroblasts mock-infected or infected (MOI = 5) with TB40/E-US28YFP or TB40/E-FLAGYFP were harvested at the indicated times and subjected to SDS-PAGE and immunoblot analysis. US28YFP, FLAGYFP, GAPDH, and molecular weight standards are indicated. (d) Fibroblasts infected (MOI = 5) with TB40/E wt, TB40/E-US28YFP or TB40/E-FLAGYFP were harvested 48 hours post-infection and visualized using the EVOS Cell Imaging Systems at 60× magnification.