Figure 1.
Mitotic errors cause chromosome instability(Au: OK?).
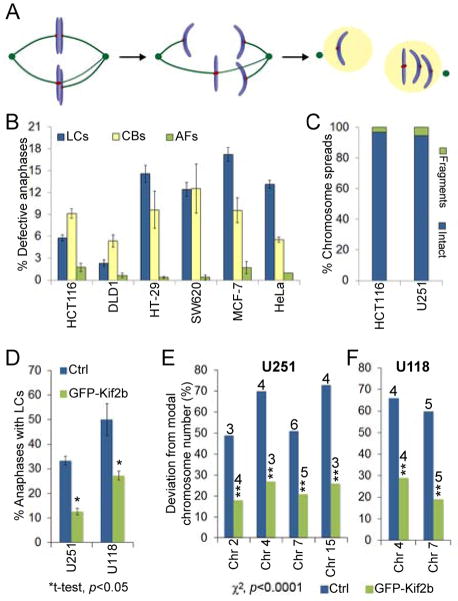
(A) Model of whole chromosome a neuploidy arising via formation of a merotelically attached (kMTs to both poles) lagging chromosome (LC) in mitosis. (B) Percent of anaphase cells with lagging chromosomes, chromatin bridges (CBs) or acentric fragments (ACs) in CIN− (HCT116 and DLD1) and CIN+ (HT-29, SW620, MCF-7, HeLa) cells. Data are represented as mean ± s.e.m., n = 448–1316 cells in 3–6 independent experiments. χ2, p<0.0001 for lagging chromosomes between CIN− and CIN+ cells.(C) Percent of chromosome spreads containing intact chromosomes or chromosome fragments in HCT116 (CIN−) and U251 (CIN+) cells, n = >300 spreads, p=0.26, χ2-test. (D) Percent of anaphase cells with lagging chromosomes in control and GFP–Kif2b over-expressing U251 and U118 cells. Data are reported as mean ± s.e.m., n = 150 cells. (E–F) Percent deviation from the modal chromosome number (noted above the bars) for the given chromosomes in a clonogenic assay (30 days) in CIN+ U251 and U118 control and GFP–Kif2b-overexpressing cells, n = 300 cells.