Figure 2. Proposed mechanism for allosteric SIRT1 activation by STACs.
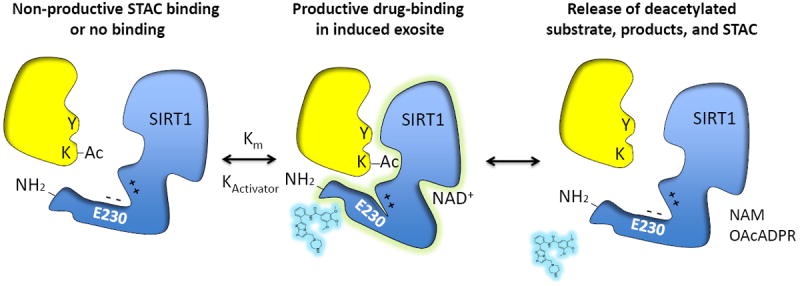
SIRT1 deacetylates target proteins (yellow) using NAD+ as a cosubstrate to generate a deacetylated product, O-acetyl-ADP ribose (OAcADPR) and nicotinamide (NAM). Biochemical and structural data favor a mechanism of direct “assisted allosteric activation” by STACs that is mediated by an N-terminal activation domain. Mutation of a conserved glutamate to lysine or alanine (E230K/A) in a structured N–terminal domain blocks activation by resveratrol and 117 synthetic STACs, arguing for a common mechanism of activation [39]. Fluorophores that were initially thought to be a requisite for activation in vitro are now known to mimic natural hydrophobic amino acids adjacent to the acetylated lysine (denoted by a Y in substrate). Binding of substrates with hydrophobic motifs near the acetyl site induce a conformational change upon binding, forming a specific exosite that allows activators to bind in a productive manner and in turn stabilize the docked substrate. Positive charges C-terminal to E230K interact with the negative charge of E230 to assist with the enzyme-STAC-substrate complex. Whether or not peptide Km corresponds to Kd is not yet known.
