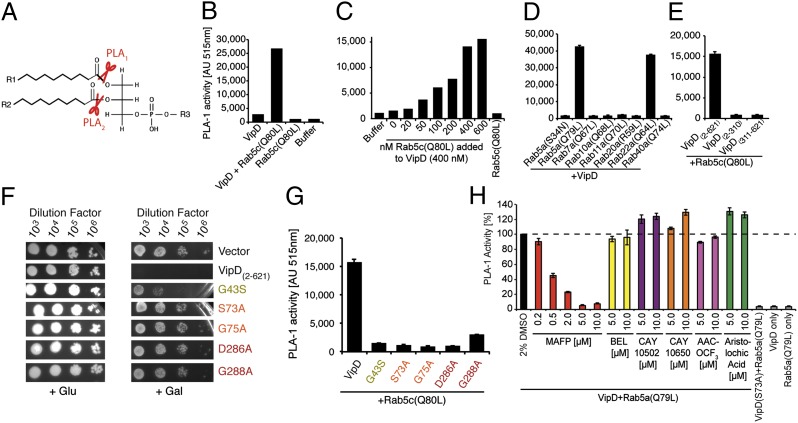
Fig. 3.
VipD exhibits PLA1 activity upon binding to active Rab5. (A) Schematic representation of PLA1 and PLA2 cleavage sites on phospholipids. (B–E) Fluorometric PLA1 assays. The substrate PED-A1 is a BODIPY FL dye-labeled glycerophosphoethanolamine, the fluorescence emission of which is dequenched upon PLA1 hydrolysis. (B) VipD exhibits PLA1 activity in the presence of Rab5c(Q80L). (C) The stimulatory effect of Rab5 on the PLA1 activity of VipD is concentration-dependent. VipD (400 nM) was incubated with increasing amounts of Rab5c(Q80L), and substrate hydrolysis was determined 40 min after incubation. (D) Active Rab22a, but none of the other Rabs tested, triggered the PLA1 activity of VipD equally efficient as Rab5a(Q79L). (E) Neither the N- nor C-terminal domain of VipD exhibited PLA1 activity in the presence of Rab5c(Q80L). (F) Substitution of catalytically relevant residues in VipD attenuated their ability to interfere with yeast growth. (G) VipD point mutants are catalytically inactive. Rab5c(Q80L) was incubated with an equimolar amount of VipD or the indicated point mutants, and substrate (PED-1A) hydrolysis was determined fluorometrically. (H) Screen for PLA2 inhibitors capable of blocking the PLA1 activity of VipD. VipD was incubated with Rab5a(Q79L) in the presence of increasing concentrations of the indicated PLA2 inhibitors. The level of uncleaved substrate 40 min after the PLA1 reaction is shown (normalized to the VipD+DMSO control).