Significance
Our establishment of a connection between the phosphatidylinositol 3-kinase (PI3K) and NRF2 pathways provides the basis for the tissue specificity of BRCA1-related cancers. Because BRCA1 is a vital component of the intracellular machinery maintaining genomic stability, BRCA1 functions as a major tumor suppressor in cells of all types. However, BRCA1-related cancers occur overwhelmingly in breasts and ovaries. Our work demonstrates that estrogen (E2) acts via the PI3K–AKT pathway to regulate NRF2 activation in BRCA1-deficient cells, resulting in the induction of antioxidant genes that protect the cell from reactive oxygen species-induced death. BRCA1-deficient cells are thus allowed to survive and may accumulate mutations, such as loss of PTEN, that perpetuate NRF2 activation. Our work supports the emerging clinical strategy of treating BRCA1-related cancers with PI3K inhibitors.
Keywords: breast cancer, reactive oxygen species, hormones, PTEN
Abstract
Mutations in the tumor suppressor BRCA1 predispose women to breast and ovarian cancers. The mechanism underlying the tissue-specific nature of BRCA1’s tumor suppression is obscure. We previously showed that the antioxidant pathway regulated by the transcription factor NRF2 is defective in BRCA1-deficient cells. Reactivation of NRF2 through silencing of its negative regulator KEAP1 permitted the survival of BRCA1-null cells. Here we show that estrogen (E2) increases the expression of NRF2-dependent antioxidant genes in various E2-responsive cell types. Like NRF2 accumulation triggered by oxidative stress, E2-induced NRF2 accumulation depends on phosphatidylinositol 3-kinase–AKT activation. Pretreatment of mammary epithelial cells (MECs) with the phosphatidylinositol 3-kinase inhibitor BKM120 abolishes the capacity of E2 to increase NRF2 protein and transcriptional activity. In vivo the survival defect of BRCA1-deficient MECs is rescued by the rise in E2 levels associated with pregnancy. Furthermore, exogenous E2 administration stimulates the growth of BRCA1-deficient mammary tumors in the fat pads of male mice. Our work elucidates the basis of the tissue specificity of BRCA1-related tumor predisposition, and explains why oophorectomy significantly reduces breast cancer risk and recurrence in women carrying BRCA1 mutations.
BRCA1 mutations promote tumor formation almost exclusively in hormone-responsive tissues such as breast and ovary (1). It has been proposed that the steroid hormone estrogen (E2) increases the survival of BRCA1-deficient cells in these tissues, favoring tumorigenesis (2). However, why BRCA1-mutated breast and ovarian epithelial cells have a survival advantage remains unclear.
E2 regulates cell differentiation, growth, and survival in a broad range of human tissues. The most abundant and potent E2 circulating in the body is 17β-estradiol. In its classical mechanism of action, E2 diffuses into the cells and binds to two nuclear E2 receptors (ERs), ERα and ERβ, which act as transcription factors and influence gene expression (3). Nonclassical mechanisms involve E2 binding to plasma membrane-associated ER proteins. An example of the latter is the phosphatidylinositol 3-kinase (PI3K) that is known to mediate E2-induced cell survival and proliferation (4). In female mice, E2 administration activates PI3K in ovarian granulosa cells (5). In vitro, E2 treatment of ER+ MCF7 human breast cancer cells stimulates the serine/threonine-protein kinase, AKT, and leads to increased glucose uptake (6) and cell cycle progression (7). E2-induced AKT activation is also involved in axonal growth and neuronal morphogenesis (8). PI3K–AKT activation in response to growth factors potentiates ERα transcriptional activity (9, 10), suggesting positive cross-talk between E2 and the PI3K–AKT pathway. Clinical studies in ER+ breast cancer patients have shown that PI3K–AKT activation underlies acquired resistance to anti-E2 or tamoxifen treatment (11). Indeed, clinicians are investigating if PI3K inhibitors can reverse anti-E2 resistance (12, 13).
Mouse studies have demonstrated that NRF2-regulated antioxidant response contributes to proliferative and mitogenic signals downstream of AKT (14). Indeed, deletion in the phosphatase and tensin homolog gene Pten potentiates the induction of antioxidant gene expression by NRF2 (14). In addition to detoxification of reactive oxygen species (ROS), AKT-regulated NRF2 controls the transcription of metabolic genes involved in the pentose phosphate pathway, de novo nucleotide synthesis, and the production of nicotinamide adenine dinucleotide phosphate (14). In a previous study (15) we showed that E2 regulates NRF2 protein levels in BRCA1-deficient mammary epithelial cells (MECs) in a manner independent of ROS induction. Given the above findings, we therefore attempted to elucidate the mechanism by which E2 regulates NRF2. Here we show that E2 controls the NRF2 antioxidant pathway through PI3K–AKT activation, and that E2s sustain the survival of BRCA1-deficient MECs in vivo. Our data suggest that E2- and PI3K–AKT-mediated control of the NRF2 antioxidant response allows both normal and malignant BRCA1-deficient cells to survive, and so is an important component of BRCA1-related tumorigenesis.
Results
E2 Induces NRF2-Regulated Antioxidant Response.
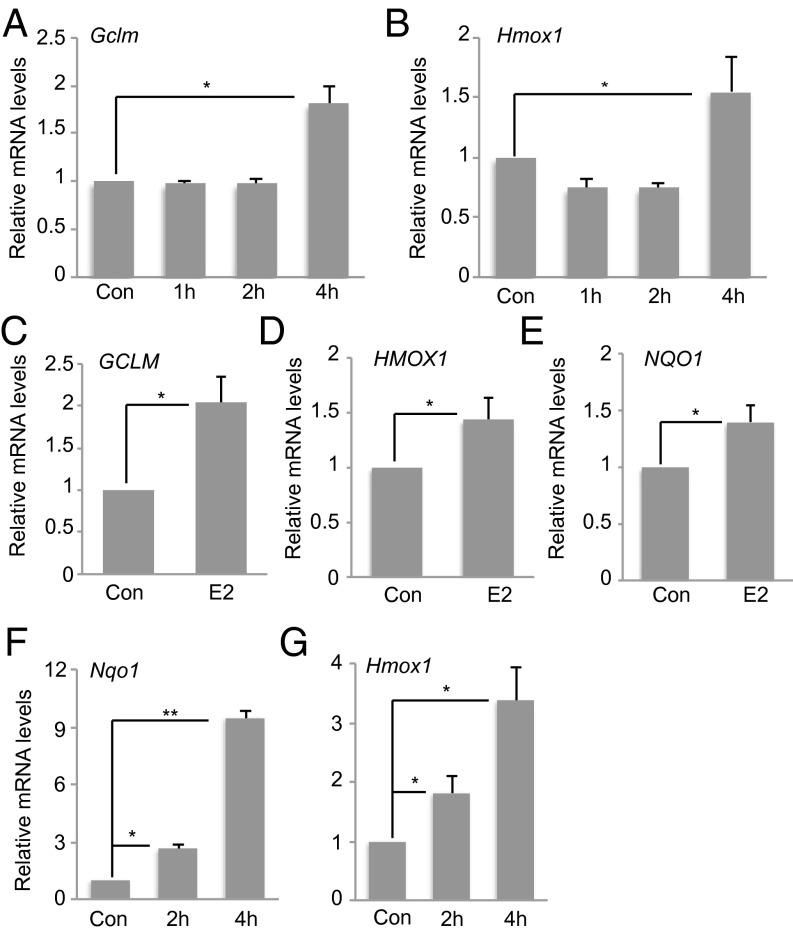
We previously reported that E2 controls NRF2 levels and activity in ER+ immortalized mouse MECs, HC11, and the ER+ human breast cancer cell line MCF7 (15). To extend these observations, we treated primary MECs (pMECs) from the mammary glands of young female mice with E2 and analyzed NRF2 activity. NRF2 target genes, glutamate-cysteine ligase, modifier subunit (GCLM) and heme oxygenase 1 (HMOX1), were up-regulated in E2-treated pMECs after 4 h (Fig. 1 A and B), confirming E2’s ability to induce the NRF2 activation.
Fig. 1.
E2 induces the expression of NRF2-regulated antioxidant genes in various cell types. Cultured pMECs were treated with 10 nM E2 for the indicated times and mRNA levels of GCLM (A) and HMOX1 (B) were assessed by RT-PCR. Con, control (no E2). Results are the mean ± SD of biological triplicates normalized to mouse rpS9 and expressed relative to the level in untreated cells (set to 1). Cultured PC-3 cells were stimulated with 10 nM E2 for 4 h and mRNA levels of GCLM (C), HMOX1 (D), and NQO1 (E) were analyzed as in A. WT mouse primary B cells were treated with LPS plus IL-4 before stimulation with 10 nM E2 for 2 or 4 h, and mRNA levels of NQO1 (F) and HMOX1 (G) were analyzed as in A. For all experiments, results are representative of at least three trials. Error bars illustrate standard error. *P < 0.05 and **P < 0.01.
In addition to affecting breast and ovary tissue, E2 regulates the growth, differentiation, and functions of nonreproductive tissues such as the prostate gland and the immune system (16). mRNA levels of GCLM, HMOX1, and NAD(P)H:quinone oxidoreductase 1 (NQO1) increased after 4-h treatment with E2 in the prostate cancer cell line PC3 (Fig. 1 C–E). Similarly, E2-treated wild-type (WT) mouse primary B cells showed elevated NQO1 and HMOX1 mRNA levels compared with controls (Fig. 1 F and G). Analysis of publicly available microarray profiling of E2-treated MCF7 cells (17) revealed the up-regulation of several other ROS-related genes (Fig. S1 and Table S1). These data indicate that E2 can trigger the NRF2-regulated antioxidant response in a wide range of cell types, and that E2 may induce multiple ROS-regulating pathways.
Oxidative Stress Induces NRF2 Through the PI3K–AKT Pathway.
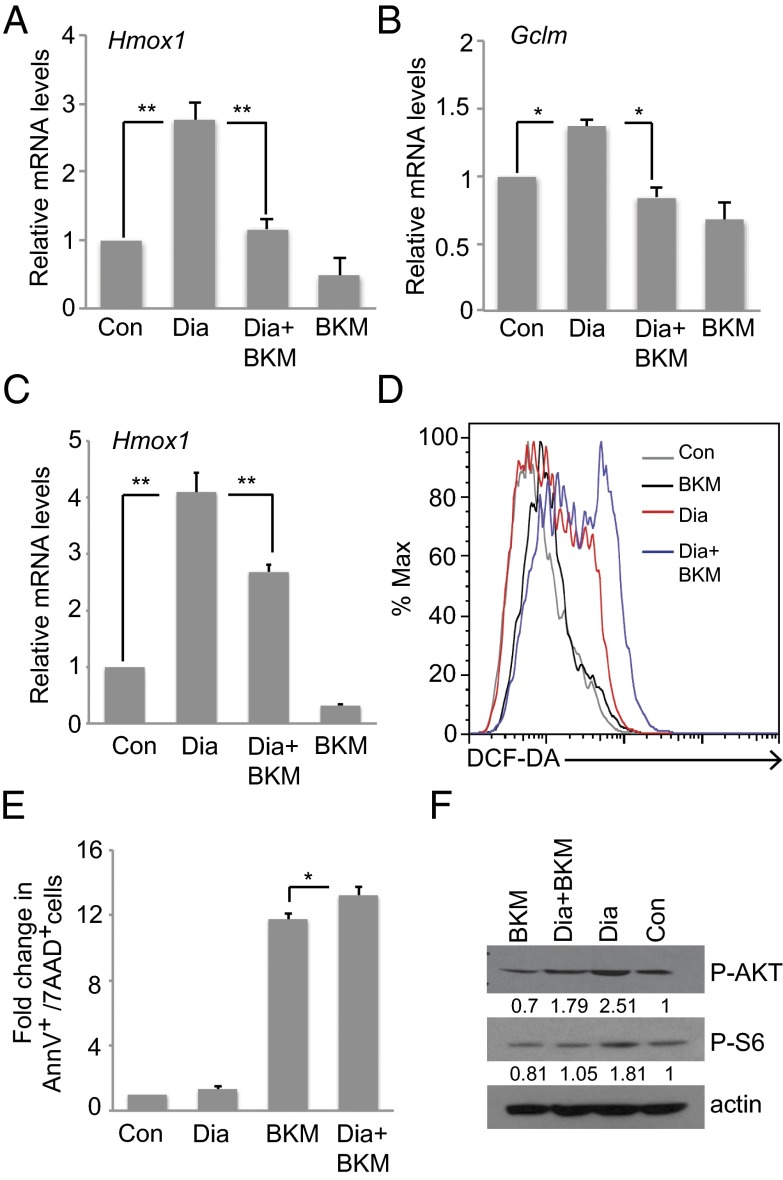
Previous work has implicated the PI3K–AKT pathway in NRF2 regulation (14). To investigate this connection upon oxidative stress, we pretreated (or not) mouse immortalized MECs COMMA-1D and HC11 cells with the PI3K inhibitor BKM120 (BKM; 1 µM), which is a highly specific class I PI3K inhibitor (18). We then treated these cells with 250 μM diamide, a thiol-oxidizing agent that oxidizes reduced glutathione and causes ROS accumulation (19). Diamide alone significantly up-regulated HMOX1 in both COMMA-1D and HC11 cells and marginally GCLM in COMMA-1D cells but in all cases BKM blocked this induction (Fig. 2 A–C). ROS levels were increased by diamide and further elevated by BKM (Fig. 2D). Apoptosis was marginally but significantly increased by BKM (Fig. 2E). Immunoblotting demonstrated that AKT and its downstream target, ribosomal protein S6, were phosphorylated after diamide, but BKM impaired this phosphorylation (Fig. 2F). These findings confirm that AKT is activated upon oxidative stress, presumably because of inactivation of the AKT negative regulator PTEN by oxidation (20–22), and that oxidative stress and PI3K signaling are linked to NRF2 activation in MECs.
Fig. 2.
Diamide induces an NRF2-regulated antioxidant response through the PI3K–AKT pathway. COMMA-1D (A and B) or HC11 (C) cells were pretreated (or not; Con) with the PI3K inhibitor BKM120 (BKM; 1 μM) before the addition of diamide (Dia; 250 μM). mRNA levels of HMOX1 (A and C) and GCLM (B) were analyzed as in Fig. 1A. (D) Intracellular ROS levels in HC11 cells treated as in C were measured as described in Materials and Methods. (E) Apoptosis was measured in HC11 cells treated as in C. Data are expressed as fold change of untreated cells (Con) and are the mean ± SD of n = 3 trials. (F) Western blot to detect phospho-AKT and phospho-S6 proteins in HC11 cells treated as in C. Actin, loading control. Error bars illustrate standard error. *P < 0.05 and **P < 0.01.
E2-Induced NRF2 Response Involves PI3K.
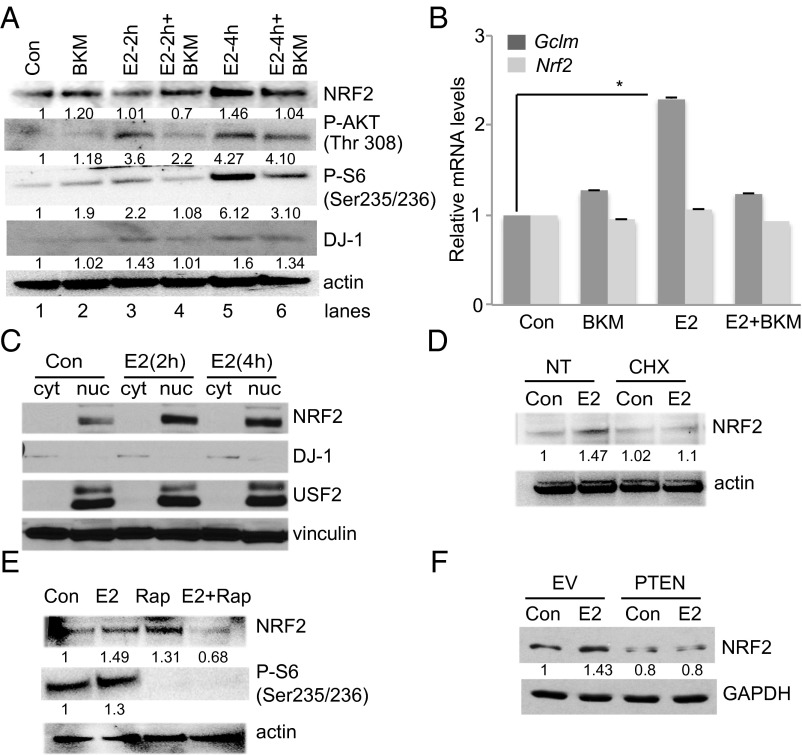
We next sought to investigate if the PI3K–AKT pathway was also involved in NRF2 regulation by E2. We E2-starved HC11 cells, pretreated them (or not) with BKM, and then stimulated them with 10 nM E2 for 2 or 4 h. NRF2 protein accumulated at 4 h post-E2 but to a much lesser extent than in BKM-pretreated cells (Fig. 3A). Accordingly, AKT was progressively phosphorylated at threonine 308 (indicating activation) (23) in response to E2, but this phosphorylation was significantly inhibited by BKM (Fig. 3A). A similar effect was observed for ribosomal protein S6 phosphorylation (Fig. 3A). Interestingly, E2 up-regulated DJ-1 in a PI3K-dependent manner (Fig. 3A). We and others have shown that DJ-1 is a cancer- and Parkinson disease-associated protein that negatively regulates PTEN, thereby promoting AKT activation (24, 25) and protects cells from oxidative stress (26, 27) by controlling mitochondrial functions (28, 29) and stabilizing NRF2 (30).
Fig. 3.
E2 triggers an NRF2-regulated antioxidant response through the PI3K–AKT pathway. (A) Western blot to detect NRF2, phospho-AKT, phospho-S6, and DJ-1 proteins in HC11 cells pretreated (or not; Con) with BKM before the addition of 10 nM E2 for 2 or 4 h, as indicated. Actin, loading control. (B) HC11 cells were treated as in A and GCLM and NRF2 mRNA levels were determined by RT-PCR as for Fig. 1A. Expression of NRF2 (C) and GREB1 (D) were analyzed in E2-treated HC11 at different time points as indicated. (E) Subcellular analysis of NRF2 protein in E2-treated HC11 after 2 and 4 h. USF2, nuclear fraction marker. DJ-1, cytoplasmic fraction marker. Vinculin, loading control. (F) Western blot to detect NRF2 protein in WAPcre-PTENf/f mammary tumor cells infected in vitro with empty vector (EV), or vector reconstituting WT PTEN (PTEN), and treated (or not; Con) with 10 nM E2 for 4 h. GAPDH, loading control. Error bars illustrate standard error. *P < 0.05.
BKM blocked NRF2 transcriptional activity, as judged by inhibition of E2-induced up-regulation of GCLM mRNA (Fig. 3B). However, NRF2 mRNA levels were not affected by either E2 or BKM (Fig. 3C). We further analyzed NRF2 mRNA expression at multiple time points of E2. Whereas the expression of GREB1, a known E2-responsive gene (31) significantly increased over time, NRF2 mRNA was not strongly induced, beside a marginal increase at 2 h (Fig. S2). Instead, we found that E2 controlled NRF2 nuclear levels by subcellular fractionation of E2-treated HC11 cells (Fig. 3C). NRF2 accumulation was due to de novo translation as assessed by pretreating E2-stimulated cells with protein synthesis inhibitor, cycloheximide (Fig. 3D). This result prompted us to investigate the role of mTOR that has been shown to promote NRF2 function (32, 33). Indeed, mTOR inhibition by rapamycin reduced NRF2 accumulation and S6 phosphorylation in E2-stimulated HC11 cells (Fig. 3E).
Recent data have shown that hyperactivated oncogenic PI3K–AKT signaling regulates NRF2 (14). We used a mouse tumor cell line derived from a primary mammary tumor spontaneously originated in a WAPcre-PTENf/f female mouse. In these mice, Cre recombinase is expressed in mammary gland secretory epithelial cells of pregnant and lactating female mice under the control of the whey acidic protein (Wap) promoter (34). We then used stable viral vector infection in vitro to reconstitute WT PTEN expression in these cells (35). Paired empty vector (EV), PTEN-deficient cells and WT PTEN-expressing tumor cells were E2-starved and stimulated with E2. Immunoblotting revealed that E2 stimulation significantly increased NRF2 protein levels in EV cells but not in cells with functional WT PTEN (Fig. 3D). These findings support our hypothesis that E2 regulates NRF2 protein levels and activity in a PI3K–mTOR-dependent manner. In B cells, E2 stimulation did not induce AKT phosphorylation and DJ-1 accumulation (Fig. S3), indicating that E2–PI3K signaling is primarily active in MECs.
E2 Sustains the Survival of BRCA1-Deficient Cells.
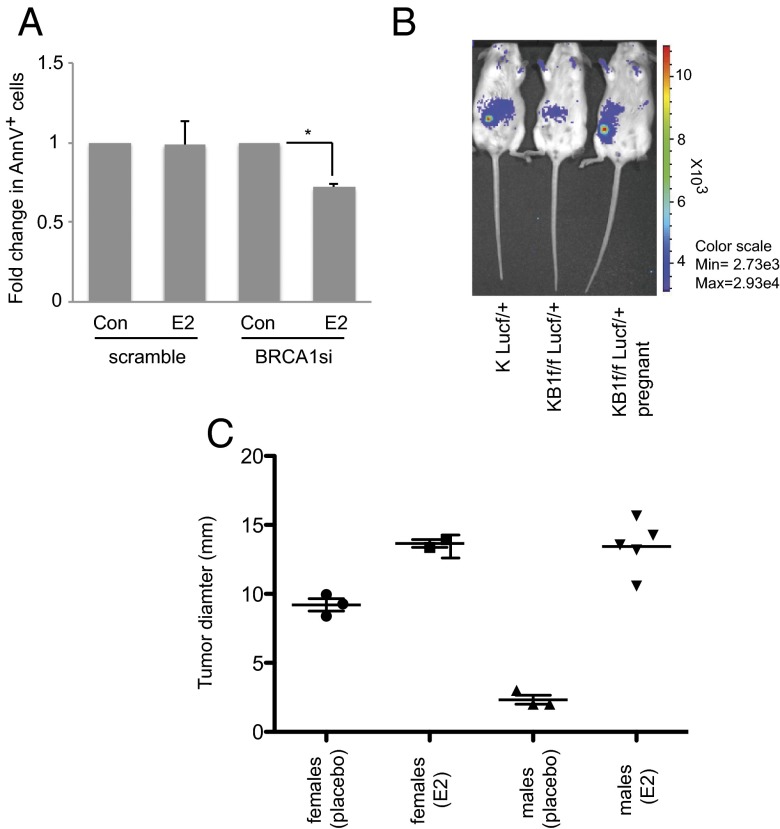
In our previous study (15) we showed that E2-induced NRF2 accumulation compensated for low NRF2 protein levels in BRCA1-null MECs, and that NRF2 activation was crucial for the survival of these cells. To determine if E2 sustains the survival of BRCA1-null cells, we first measured apoptosis in E2-stimulated HC11 cells transfected with either nontargeting siRNA (scrambled) or mouse BRCA1-specific siRNA (BRCA1si). AnnexinV/7-AAD staining revealed that 4-h E2 treatment rescued early apoptosis (AnnexinV+ cells) in BRCA1-null cells (Fig. 4A). We then isolated pMECs from K14cre BRCA1f/fLUCf/f female reporter mice (15) and implanted them in the precleared fat pads of normal young females. At 8 wk posttransplantation, luciferase signals of mammary outgrowths were scored by in vivo imaging system (IVIS). BRCA1-null cells failed to reconstitute a functional mammary gland in nulliparous recipients but gave rise to luciferase-positive outgrowths in pregnant females (Fig. 4B). This suggests that the E2-rich environment of a pregnant mouse sustained survival of BRCA1-deficient pMECs.
Fig. 4.
E2 promotes the survival of both normal and malignant BRCA1-deficient MECs. (A) HC11 cells were transfected with scrambled or BRCA1-specific (BRCA1si) siRNA oligomers as described in Materials and Methods. At 24 h posttransfection, cells were stimulated (or not; Con) with 10 nM E2 for 4 h and apoptosis was measured by AnnexinV staining. Results are mean ± SD (n = 3). *P < 0.05. (B) pMECs isolated from KLucf/+ or KB1f/fLucf/+ donor mice were transplanted into 21-wk-old recipient female mice (n = 3 per group). At 1 wk posttransplantation, n = 3 female mice that had received KB1f/fLucf/+ pMECs were bred to a WT male and became pregnant. At 8 wk posttransplantation, in vivo luciferase activity in outgrowths derived from the implanted pMECs was determined by IVIS. (C) Male and female mice were injected in fat pads with KBP cells (n = 3–5 per group) and injected s.c. in the neck with a pellet containing vehicle or 0.72 mg E2. Tumor cell growth was monitored for 30 d and palpable tumor diameters were measured. Datapoints are single tumors in individual mice. Horizontal lines are geometric means ± SD.
Although BRCA1-mutated tumors are considered ER−, mouse and human studies have shown that these tumors are still responsive to E2 (36–39). To investigate this response, we injected mammary tumor cells from K14cre BRCA1f/fp53f/f (KBP) mice (40) into the fat pads of normal female and male mice. Pellets containing either vehicle or .72 mg E2 were s.c. implanted in the necks of these animals and tumor cell growth was monitored. In vehicle-treated males, KBP tumor cells grew poorly compared with the same cells injected into females (Fig. 4C). However, E2 treatment increased KBP growth in female mice and rescued KBP survival in males (Fig. 4C). Our results show that E2 has a powerful positive effect on the survival of both BRCA1-deficient normal and tumor cells.
Discussion
Our study has delineated a strong relationship between PI3K signaling and the NRF2-regulated antioxidant response in MECs. These two pathways are interconnected during responses to either oxidative stress or E2 stimulation. In both cases, we found that PI3K activation is required for efficient NRF2 target gene transcription (Figs. 2 and 3). Upon oxidative stress, failure to activate NRF2 due to PI3K inhibition resulted in ROS accumulation and increased cell death (Fig. 2). Upon E2 treatment, NRF2 protein accumulated and drove gene expression contributing to PI3K-driven survival (Fig. 3). E2 controls NRF2 translation (Fig. 3D). Interestingly, activation of the mammalian target of rapamycin (mTOR) is involved in E2-induced NRF2 regulation (Fig. 3E).
Our study indicates that ROS control mediated by the NRF2 antioxidant response contributes to AKT-stimulated mitogenic and survival programs. It is now clear that ROS are not simply by-stander products of metabolic reactions but exert myriad functions with important implications for tumor development (41). Antioxidant pathways can promote tumorigenesis by supporting tumor cell survival (42, 43). Therefore, tumor cells can be eliminated by targeting their mechanism of antioxidant adaptation (44).
Our results indicate that NRF2 has an oncogenic role in BRCA1-associated tumorigenesis. We previously showed that BRCA1 interacts with NRF2, that BRCA1-deficient cells accumulate ROS as a result of a defective NRF2 response, and that NRF2 reactivation rescues cell survival (15). E2 is a key component of this survival pathway, because E2-regulated NRF2 compensates for the lack of an antioxidant response in the absence of BRCA1. Here we show that the PI3K–AKT pathway is another crucial step in NRF2 regulation, and that it is PI3K activation that mediates the capacity of E2 to trigger the NRF2 antioxidant response. Indeed, in response to E2, mouse mammary tumor cells lacking PTEN show both hyperactivated AKT and stimulated NRF2. Significantly, PTEN mutations are very frequent in BRCA1-mutated tumors (45), and the resulting PI3K–AKT hyperactivation sustains the growth of these cancers (46). For this reason, PI3K inhibitors are currently undergoing preclinical and clinical study as promising therapeutic agents for BRCA1-mutated tumors (47, 48).
Accumulating evidence suggests that E2 plays a significant role in the etiology and treatment of BRCA1-related cancers despite the lack of ER expression by these tumors. First, BRCA1-related tumors are largely restricted to hormone-responsive tissues, such as those of the breast and ovary (49). Second, oophorectomy significantly reduces breast cancer risk and recurrence in carriers of BRCA1 mutations (36–38). Third, proliferation of premalignant BRCA1-deficient cells in a BRCA1 knockout mouse model is E2-dependent (39). Lastly, ER is a substrate of BRCA1 ubiquitin ligase activity, suggesting that ER accumulation may drive proliferation of breast and ovary tissues in the absence of BRCA1 (50).
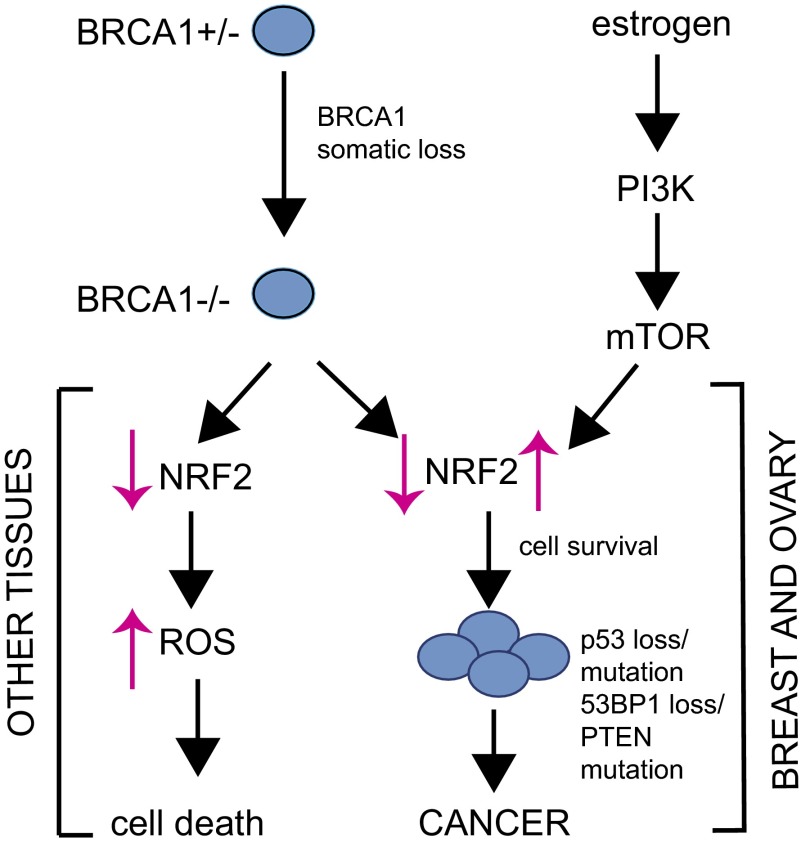
Our work in the present study has bolstered these findings by showing that E2, NRF2, PI3K, and BRCA1 are all intricately linked during BRCA1-related tumorigenesis. Our results suggest the following model (Fig. 5): where BRCA1 somatic loss in heterozygous carriers of BRCA1 mutations has differential effects depending on the tissue. In tissues that do not provide an E2-rich environment, BRCA1 deficiency impairs NRF2 antioxidant signaling, leading to increased ROSs that kill BRCA1-deficient cells. However, in breasts and ovaries, E2 protects BRCA1-deficient cells from oxidative stress-induced death by activating NRF2 via a mechanism that depends on PI3K–AKT. If a BRCA1-deficient cell loses PTEN function, the PI3K–AKT pathway may be further stimulated and thus reinforce E2-mediated NRF2 signaling. Mitogenic and antioxidant pathways acting downstream of AKT, coupled with the genomic instability caused by a lack of BRCA1-mediated DNA repair, will eventually result in malignant transformation of BRCA1-null cells.
Fig. 5.
Model of the role of NRF2 regulation in BRCA1-associated tumorigenesis. See detailed description in the Discussion.
In conclusion, we have shown that E2 regulates the NRF2 antioxidant response via a PI3K–AKT-regulated signaling pathway that may involve DJ-1 and mTOR (Fig. 3). We have also demonstrated that E2 is important for controlling the survival of both normal and malignant BRCA1-deficient cells (Fig. 4). Our work provides a rationale for the use of PI3K inhibitors in the treatment of BRCA1-mutated tumors, because these inhibitors should block NRF2 activity in BRCA1-deficient cells, thereby increasing ROS and promoting cell death.
Materials and Methods
Mice.
K14cre transgenic mice and p53 conditional knockout mice were from A. Berns [Netherlands Cancer Institute (NKI), Amsterdam] (51). Brca1 conditional knockout mice were from J. Jonkers (NKI) (40). Luciferase-expressing transgenic mice were from the Jackson Laboratory (52). For mammary gland studies and in vivo E2 studies, mice were treated as described (15, 53). For xenograft studies, mammary tumor cells were implanted into the fat pads of WT mice. All mouse protocols were approved by the Animal Care and Use Committee of the University Health Network (Toronto).
Mouse pMECs and Fat Pad Transplantation.
Mouse pMECs obtained by dissociating the left and right inguinal mammary glands of KLucf/+ or KB1f/fLucf/+ female mice were maintained in culture, processed for transplantation, and tracked by IVIS as described (15). Shortly after transplantation, three recipient females were bred with a male and became pregnant. Luciferase signals were assessed as soon as pregnancy become evident at ∼10 d postmating.
Tumor Xenografts.
Cells were isolated from a mammary tumor originating in a K14cre BRCA1f/fp53f/f female mouse after disruption of tumor tissue with collagenase. Cells were maintained in DMEM/F12 medium containing 10% FBS, L-glutamine, 1 μg/mL hydrocortisone (Sigma), 5 μg/mL insulin (Sigma), and 5 ng/mL epidermal growth factor (Sigma) as pMECs in a humidified tissue culture incubator with 3% oxygen. After a few weeks in culture, tumor cells became immortalized (KBP cell line). KBP cells (1.5 × 105) were injected into the fat pads of WT mice and a pellet containing either vehicle or E2 (0.72 mg, 90-d release from Innovative Research of America) was simultaneously introduced s.c. into the neck. Tumor growth was monitored daily and palpable tumor diameters were measured with a caliper.
Cell Lines and Culture Conditions.
COMMA-1D, HC11, and MC7 cells were obtained and maintained as described (15, 53). Human PC-3 cells were from American Tissue Culture Collection. WAPcre-PTEN cells were obtained and treated as described (35). Primary B cells were isolated from WT FVB mice with a Miltenyi Biotech B-cell isolation kit and immediately processed for E2 stimulation in RPM1, 10% FBS, and l-glutamine. IgM Ig was used at 20 μg/mL.
For E2 experiments, MCF7, HC11, B cells, or pMECs were maintained for 2–3 d in their respective culture media (phenol red-free) supplemented with 10% charcoal-filtered FBS. Ten nanomolars of 17β-estradiol (E2; Sigma) were added to E2-starved cultures for 2 or 4 h. Rapamycin (100 nM) and BKM (1 μM) [gifts from V. Stambolic (Ontario Cancer Institute, Toronto)] were added 1 h prior E2 and kept during E2 treatment. Cycloheximide (25 μM) was added 30 min before E2 and kept in the media. To induce oxidative stress, cells were treated for 2 h with 250 μM diamide (Sigma).
RT-PCR.
RNA was isolated and processed by quantitative RT-PCR as described (15). Data were normalized to mouse ribosomal rpS9 and human ribosomal rps18.
AnnexinV–7-AAD Staining.
Cells were stained with FITC-conjugated AnnexinV plus 7-AAD for 15 min at room temperature in 1× AnnexinV binding buffer (10 mM Hepes⋅sodium hydroxide, pH 7.4, 140 mM NaCl, 2.5 mM CaCl2). All reagents were from BD Biosciences. Cells were analyzed by BD FACSCalibur flow cytometer immediately after staining.
Intracellular ROS Staining.
Cells were incubated with 300 nM dichloro-dihydro-fluorescein diacetate (DCF-DA) (C6827; Invitrogen) for 10 min at 37 °C. After two washes in PBS-1×, DCF-DA fluorescence was analyzed using a FACS Canto flow cytometer (BD Biosciences) and FlowJo software. Mean fluorescence values (FL-1 for DCF-DA) were displayed as histogram overlays using the %Max option, which scaled each population curve to mode equal to 100% on the y axis and log10 FL-1 (DCF-DA) fluorescence intensity on the x axis.
siRNA Transfection.
HC11 cells (105) in six-well plates were transfected with 100 pmol of nontargeting scrambled siRNA (d-001810–10; Dharmacon) or siRNA targeting mouse Brca1 (l-040545-00; Dharmacon) using Lipofectamine 2000. Transfection medium was removed after 5 h and cells were cultured for an additional 48 h in E2-starving media before experiments.
Subcellular Fractionation.
HC11 cells were E2-starved and stimulated with E2 for 2 and 4 h. Cellular cytoplasmic and nuclear fractions were obtained by using NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo-Scientific) according to the manufacturer’s instructions. Twenty micrograms of lysate from each fraction were processed by Western blotting.
Western Blotting.
Cells were lysed in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris, pH 8.0) on ice for 15 min, sonicated, and centrifuged at 13,000 × g for 15 min. The protein content of supernatants was measured by BCA assay (Pierce). Thirty micrograms of protein samples were fractionated by reducing 4–12% SDS/PAGE, transferred to PVDF membranes, and incubated overnight with the following primary antibodies: anti-NRF2 (kindly provided by E. Schmidt, Montana State University), anti–pAKT-T308 (catalog no. 2965; Cell Signaling), anti–pAKT-S437 (catalog no. 4058; Cell Signaling), anti–pS6-S235/236 (catalog no. 2215; Cell Signaling), and anti–β-actin (Abcam) and anti-vinculin (Abcam). Antibody against DJ-1 was generated as described (54). Primary antibodies were detected using HRP-conjugated secondary antibodies to rabbit IgG (GE Healthcare). Immunocomplexes were visualized using enhanced luminol-based chemiluminescent (GE Healthcare). When required, immunoblots were quantified by ImageJ 1.45 software (55).
Gene Microarray.
To examine the alterations to the transcription of genes encoding oxidative stress pathway-related proteins, we constructed an expanded gene set by aggregating previously published experimentally and manually defined datasets. Using the Broad Institute Molecular Signatures Database (www.broadinstitute.org/gsea/msigdb/index.jsp), we conducted searches using the term “oxidative” and combined the following gene sets: CHUANG_OXIDATIVE_STRESS_RESPONSE_DN, CHUANG_OXIDATIVE_STRESS_RESPONSE_UP, KEGG_OXIDATIVE_PHOSPHORYLATION, WEIGEL_OXIDATIVE_STRESS_RESPONSE, WEIGEL_OXIDATIVE_STRESS_BY_TBH_AND_H2O2, WEIGEL_OXIDATIVE_STRESS_BY_HNE_AND_TBH, WEIGEL_OXIDATIVE_STRESS_BY_HNE_AND_H2O2, and RESPONSE_TO_OXIDATIVE_STRESS.
Redundant entries were removed from the resulting list and differential expression of the remaining genes was investigated in gene expression data obtained from the National Center for Biotechnology Information Gene Expression Omnibus database (accession no. GSE11324). The raw probe intensity (.CEL) files were downloaded and processed with a custom chip definition file (.CDF) to update the probe annotations (56). Data were background-corrected, normalized, and summarized using the robust multiarray average algorithm (57) as implemented in the R statistical language (http://r-project.org). The difference in gene expression level between untreated and E2-treated cells was assessed using the Linea Models for Microarray Data (LIMMA) method (58) implemented using the LIMMA package in the R language. Genes for which the false discovery rate was below 5% were considered differentially expressed at a statistically significant level. The significant Gene Ontology biological process clusters were obtained with the Web-based tool DAVID (59).
Statistical Analyses.
Unless otherwise indicated, results were reported as the mean ± SD. The two-sided independent Student t test without equal variance assumption was used to determine P values *P < 0.05 and **P < 0.01.
Supplementary Material
Acknowledgments
We thank M. Saunders for scientific editing. This research was supported by grants from the Canadian Institutes of Health Research (to T.W.M.) and a grant from the Ontario Ministry of Health and Long Term Care (to T.W.M.).
Footnotes
The authors declare no conflict of interest.
See Commentary on page 4353.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1324136111/-/DCSupplemental.
References
- 1.Valentin MD, da Silva SD, Privat M, Alaoui-Jamali M, Bignon YJ. Molecular insights on basal-like breast cancer. Breast Cancer Res Treat. 2012;134(1):21–30. doi: 10.1007/s10549-011-1934-z. [DOI] [PubMed] [Google Scholar]
- 2.Elledge SJ, Amon A. The BRCA1 suppressor hypothesis: An explanation for the tissue-specific tumor development in BRCA1 patients. Cancer Cell. 2002;1(2):129–132. doi: 10.1016/s1535-6108(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 3.Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116(3):561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renoir JM, Marsaud V, Lazennec G. Estrogen receptor signaling as a target for novel breast cancer therapeutics. Biochem Pharmacol. 2013;85(4):449–465. doi: 10.1016/j.bcp.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 5.Li Q, et al. Phosphoinositide 3-kinase p110δ mediates estrogen- and FSH-stimulated ovarian follicle growth. Mol Endocrinol. 2013;27(9):1468–1482. doi: 10.1210/me.2013-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garrido P, Morán J, Alonso A, González S, González C. 17β-estradiol activates glucose uptake via GLUT4 translocation and PI3K/Akt signaling pathway in MCF-7 cells. Endocrinology. 2013;154(6):1979–1989. doi: 10.1210/en.2012-1558. [DOI] [PubMed] [Google Scholar]
- 7.Gaben AM, Sabbah M, Redeuilh G, Bedin M, Mester J. Ligand-free estrogen receptor activity complements IGF1R to induce the proliferation of the MCF-7 breast cancer cells. BMC Cancer. 2012;12:291. doi: 10.1186/1471-2407-12-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varea O, Escoll M, Diez H, Garrido JJ, Wandosell F. Oestradiol signalling through the Akt-mTORC1-S6K1. Biochim Biophys Acta. 2013;1833(5):1052–1064. doi: 10.1016/j.bbamcr.2012.12.019. [DOI] [PubMed] [Google Scholar]
- 9.Martin MB, et al. A role for Akt in mediating the estrogenic functions of epidermal growth factor and insulin-like growth factor I. Endocrinology. 2000;141(12):4503–4511. doi: 10.1210/endo.141.12.7836. [DOI] [PubMed] [Google Scholar]
- 10.Bratton MR, et al. Regulation of ERalpha-mediated transcription of Bcl-2 by PI3K-AKT crosstalk: Implications for breast cancer cell survival. Int J Oncol. 2010;37(3):541–550. doi: 10.3892/ijo_00000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Provenzano A, Kurian S, Abraham J. Overcoming endocrine resistance in breast cancer: Role of the PI3K and the mTOR pathways. Expert Rev Anticancer Ther. 2013;13(2):143–147. doi: 10.1586/era.12.173. [DOI] [PubMed] [Google Scholar]
- 12.Fox EM, Arteaga CL, Miller TW. Abrogating endocrine resistance by targeting ERα and PI3K in breast cancer. Front Oncol. 2012;2:145. doi: 10.3389/fonc.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miller TW, Balko JM, Arteaga CL. Phosphatidylinositol 3-kinase and antiestrogen resistance in breast cancer. J Clin Oncol. 2011;29(33):4452–4461. doi: 10.1200/JCO.2010.34.4879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitsuishi Y, et al. Nrf2 redirects glucose and glutamine into anabolic pathways in metabolic reprogramming. Cancer Cell. 2012;22(1):66–79. doi: 10.1016/j.ccr.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Gorrini C, et al. BRCA1 interacts with Nrf2 to regulate antioxidant signaling and cell survival. J Exp Med. 2013;210(8):1529–1544. doi: 10.1084/jem.20121337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diel P. Tissue-specific estrogenic response and molecular mechanisms. Toxicol Lett. 2002;127(1-3):217–224. doi: 10.1016/s0378-4274(01)00503-3. [DOI] [PubMed] [Google Scholar]
- 17.Carroll JS, et al. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38(11):1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 18.Maira SM, et al. Identification and characterization of NVP-BKM120, an orally available pan-class I PI3-kinase inhibitor. Mol Cancer Ther. 2012;11(2):317–328. doi: 10.1158/1535-7163.MCT-11-0474. [DOI] [PubMed] [Google Scholar]
- 19.Kosower NS, Kosower EM, Wertheim B, Correa WS. Diamide, a new reagent for the intracellular oxidation of glutathione to the disulfide. Biochem Biophys Res Commun. 1969;37(4):593–596. doi: 10.1016/0006-291x(69)90850-x. [DOI] [PubMed] [Google Scholar]
- 20.Chetram MA, et al. ROS-mediated activation of AKT induces apoptosis via pVHL in prostate cancer cells. Mol Cell Biochem. 2013;376(1-2):63–71. doi: 10.1007/s11010-012-1549-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Covey TM, Edes K, Coombs GS, Virshup DM, Fitzpatrick FA. Alkylation of the tumor suppressor PTEN activates Akt and β-catenin signaling: A mechanism linking inflammation and oxidative stress with cancer. PLoS ONE. 2010;5(10):e13545. doi: 10.1371/journal.pone.0013545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leslie NR, et al. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22(20):5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Scheid MP, Marignani PA, Woodgett JR. Multiple phosphoinositide 3-kinase-dependent steps in activation of protein kinase B. Mol Cell Biol. 2002;22(17):6247–6260. doi: 10.1128/MCB.22.17.6247-6260.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang Y, et al. Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc Natl Acad Sci USA. 2005;102(38):13670–13675. doi: 10.1073/pnas.0504610102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim RH, et al. DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell. 2005;7(3):263–273. doi: 10.1016/j.ccr.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 26.Billia F, et al. Parkinson-susceptibility gene DJ-1/PARK7 protects the murine heart from oxidative damage in vivo. Proc Natl Acad Sci USA. 2013;110(15):6085–6090. doi: 10.1073/pnas.1303444110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shadrach KG, Rayborn ME, Hollyfield JG, Bonilha VL. DJ-1-dependent regulation of oxidative stress in the retinal pigment epithelium (RPE) PLoS ONE. 2013;8(7):e67983. doi: 10.1371/journal.pone.0067983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Irrcher I, et al. Loss of the Parkinson’s disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum Mol Genet. 2010;19(19):3734–3746. doi: 10.1093/hmg/ddq288. [DOI] [PubMed] [Google Scholar]
- 29.Joselin AP, et al. ROS-dependent regulation of Parkin and DJ-1 localization during oxidative stress in neurons. Hum Mol Genet. 2012;21(22):4888–4903. doi: 10.1093/hmg/dds325. [DOI] [PubMed] [Google Scholar]
- 30.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson’s disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci USA. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh MG, Thompson DA, Weigel RJ. PDZK1 and GREB1 are estrogen-regulated genes expressed in hormone-responsive breast cancer. Cancer Res. 2000;60(22):6367–6375. [PubMed] [Google Scholar]
- 32.Bray K, et al. Autophagy suppresses RIP kinase-dependent necrosis enabling survival to mTOR inhibition. PLoS ONE. 2012;7(7):e41831. doi: 10.1371/journal.pone.0041831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shibata T, et al. Global downstream pathway analysis reveals a dependence of oncogenic NF-E2-related factor 2 mutation on the mTOR growth signaling pathway. Cancer Res. 2010;70(22):9095–9105. doi: 10.1158/0008-5472.CAN-10-0384. [DOI] [PubMed] [Google Scholar]
- 34.Wagner KU, et al. Cre-mediated gene deletion in the mammary gland. Nucleic Acids Res. 1997;25(21):4323–4330. doi: 10.1093/nar/25.21.4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bassi C, et al. Nuclear PTEN controls DNA repair and sensitivity to genotoxic stress. Science. 2013;341(6144):395–399. doi: 10.1126/science.1236188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Domchek SM, et al. Occult ovarian cancers identified at risk-reducing salpingo-oophorectomy in a prospective cohort of BRCA1/2 mutation carriers. Breast Cancer Res Treat. 2010;124(1):195–203. doi: 10.1007/s10549-010-0799-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Domchek SM, et al. Breast cancer risks in individuals testing negative for a known family mutation in BRCA1 or BRCA2. Breast Cancer Res Treat. 2010;119(2):409–414. doi: 10.1007/s10549-009-0611-y. [DOI] [PubMed] [Google Scholar]
- 38.Evans DG, et al. Contralateral mastectomy improves survival in women with BRCA1/2-associated breast cancer. Breast Cancer Res Treat. 2013;140(1):135–142. doi: 10.1007/s10549-013-2583-1. [DOI] [PubMed] [Google Scholar]
- 39.Li W, Xiao C, Vonderhaar BK, Deng CX. A role of estrogen/ERalpha signaling in BRCA1-associated tissue-specific tumor formation. Oncogene. 2007;26(51):7204–7212. doi: 10.1038/sj.onc.1210527. [DOI] [PubMed] [Google Scholar]
- 40.Liu X, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci USA. 2007;104(29):12111–12116. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11(2):85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 42.DeNicola GM, et al. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature. 2011;475(7354):106–109. doi: 10.1038/nature10189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schafer ZT, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461(7260):109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gorrini C, Harris IS, Mak TW. Modulation of oxidative stress as an anticancer strategy. Nat Rev Drug Discov. 2013;12(12):931–947. doi: 10.1038/nrd4002. [DOI] [PubMed] [Google Scholar]
- 45.Saal LH, et al. Recurrent gross mutations of the PTEN tumor suppressor gene in breast cancers with deficient DSB repair. Nat Genet. 2008;40(1):102–107. doi: 10.1038/ng.2007.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foulkes WD. BRCA1—sowing the seeds crooked in the furrow. Nat Genet. 2008;40(1):8–9. doi: 10.1038/ng0108-8. [DOI] [PubMed] [Google Scholar]
- 47.Juvekar A, et al. Combining a PI3K inhibitor with a PARP inhibitor provides an effective therapy for BRCA1-related breast cancer. Cancer Discov. 2012;2(11):1048–1063. doi: 10.1158/2159-8290.CD-11-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ibrahim YH, et al. PI3K inhibition impairs BRCA1/2 expression and sensitizes BRCA-proficient triple-negative breast cancer to PARP inhibition. Cancer Discov. 2012;2(11):1036–1047. doi: 10.1158/2159-8290.CD-11-0348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hilakivi-Clarke L. Estrogens, BRCA1, and breast cancer. Cancer Res. 2000;60(18):4993–5001. [PubMed] [Google Scholar]
- 50.Eakin CM, Maccoss MJ, Finney GL, Klevit RE. Estrogen receptor alpha is a putative substrate for the BRCA1 ubiquitin ligase. Proc Natl Acad Sci USA. 2007;104(14):5794–5799. doi: 10.1073/pnas.0610887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jonkers J, et al. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat Genet. 2001;29(4):418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 52.Safran M, et al. Mouse reporter strain for noninvasive bioluminescent imaging of cells that have undergone Cre-mediated recombination. Mol Imaging. 2003;2(4):297–302. doi: 10.1162/15353500200303154. [DOI] [PubMed] [Google Scholar]
- 53.Joshi PA, et al. Progesterone induces adult mammary stem cell expansion. Nature. 2010;465(7299):803–807. doi: 10.1038/nature09091. [DOI] [PubMed] [Google Scholar]
- 54.Li Y, et al. Dilated cardiomyopathy and neonatal lethality in mutant mice lacking manganese superoxide dismutase. Nat Genet. 1995;11(4):376–381. doi: 10.1038/ng1295-376. [DOI] [PubMed] [Google Scholar]
- 55.Schneider CA, Rasband WS, Eliceiri KW. NIH Image to ImageJ: 25 years of image analysis. Nature Methods. 2012 doi: 10.1038/nmeth.2089. 10.1038/nmeth.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dai M, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33(20):e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 58.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21(9):2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 59.Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.