Figure 5.
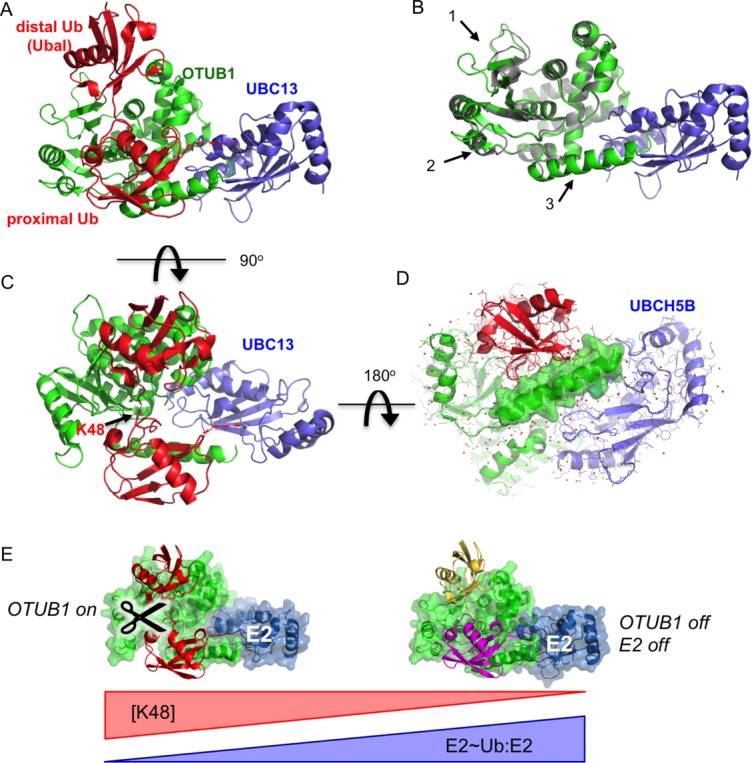
Cross-regulation by OTUB1 and E2 enzymes. A: Structure of OTUB1 bound to ubiquitin aldehyde (Ubal) and a UBC13∼Ub conjugate. OTUB1 in this figure is a hybrid containing the catalytic domain of C. elegans OTUB1 and the N-terminal helix of human OTUB160 (4DHZ). B: Superposition of OTUB1 in the presence (green) and absence (gray) of bound ubiquitin. UBC13 shown in blue. Arrows indicate conformational changes in the globular domain (1 and 2) and formation of an N-terminal ubiquitin-binding helix (3) (4DHZ and 4DHI). C: Binding of ubiquitin to distal (top) and proximal (bottom) sites in OTUB1 mimics K48-linked diubiquitin substrate. View of complex in (A) rotated by 90° shows juxtaposition of K48 of proximal ubiquitin and the C-terminus of the distal ubiquitin. D: Structure of OTUB1-Ubal bound to UBCH5B∼Ub59 (4LDT). View of complex is rotated 180° relative to view of OTUB1-Ubal-UBC13∼Ub complex shown in (C). Surface rendering of OTUB1 C-terminal ubiquitin-binding helix illustrates how this helix, which forms only in OTUB1-E2 complexes containing two bound ubiquitins, forms contacts with both UBCH5B and the proximal ubiquitin. E: Model for regulation of OTUB1 activity in response to changes in ratio of uncharged to charged E2 (E2:E2∼Ub) as well as the concentration of K48-linked polyubiquitin. At low levels of E2 charging (left), E2 binding to OTUB1 stimulates cleavage of K48-linked polyubiquitin. A high proportion of charged E2∼Ub (right), the repressed complex will form between the E2∼Ub, OTUB1 and a free ubiquitin monomer (yellow), which blocks both OTUB1 DUB activity and the ability of the E2 to conjugate ubiquitin to substrate.
