Figure 3.
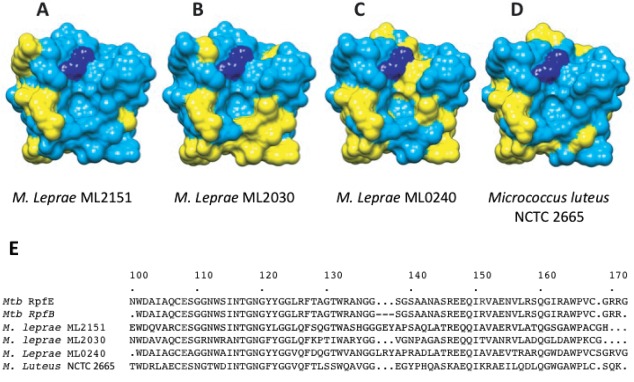
Sequence variation for RpfE and Mycobacterium leprae homologs. Conserved (blue) and variable residues (yellow) in RpfE and ML2151, ML2030, and ML0240, respectively, (A), (B), and (C). Sequence differences are prominent in the predicted peptide-binding groove to the right of the catalytic glutamate (dark blue) and the predicted carbohydrate-binding surface below the active-site pocket. D: Sequence variation between RpfE and the most similar protein in Micrococcus luteus (51% sequence identity) shows homologies in a more distantly related Rpf. E: Sequence aligment of Mtb RpfE and RpfB catalytic domain with M. leprae and M. luteus Rpfs.
