Figure 1.
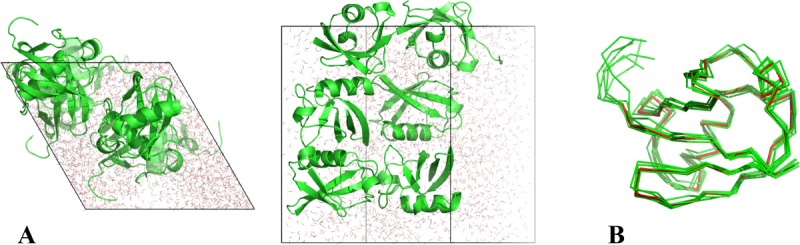
(A) The snapshot from erMD simulation of ubiquitin showing periodic-boundary box (corresponding to the single crystal unit cell, 1U). The unit cell with the primitive trigonal space group P3221 is based on the crystallographic structure 3ONS. The reported dimensions of the cell, a=b and c, are all increased by a factor 1.016 to account for thermal expansion of the protein crystal on transition from 100 K (temperature at which 3ONS was solved) to 301 K (temperature at which ssNMR data were taken).27 Shown are the top view and side view of the unit cell. The MD trajectory was recorded with k0 kcal mol−1 Å−2; the displayed snapshot represents the time point 150 ns. The areas with apparent low water density arise from the periodic-boundary images of ubiquitin molecules. (B) Six ubiquitin molecules from the MD frame, panel A, superimposed according to Eq. (2.1) (green Cα traces). Also shown is the crystallographic structure 3ONS centered according to Eq. (2.2) (red Cα trace). Such superpositions are used to calculate the instantaneous value of Urestraint, Eq. (1). Since protein molecules are superimposed via the symmetry transformations rather than least-square fitting, Urestraint proves to be sensitive to small reorientations of proteins in the simulated unit cell.
