Linkage disequilibrium (LD) and confounding are two widely discussed concepts in Genetics and in Epidemiology, yet their relationship has received only intuitive or no considerations. Taken in the narrow sense, LD refers to the nonrandom association of alleles at two or more linked genetic loci. The degree to which these alleles are associated is the basis behind genetic association studies whereby the genetic variation across target chromosomal intervals or over the entire genome is captured by a set of representative genetic markers (tagging markers). The denomination of ‘linkage' is somehow misleading because LD is a special case of the more general gametic phase disequilibrium (GPD),1 which also occurs between variants at the mitochondrial and nuclear loci in the form of cytonuclear disequilibrium.2
Confounding occurs when an extraneous factor, or a set of factors, can at least partially explain an apparent association or a lack of an apparent association between a risk factor and the outcome. In the former case, the confounding variable, or confounder, causes the association to appear, whereas in the second case the confounder masks a real association.3 The classical example is the apparent increase of risk for heart disease with alcohol consumption, an association that might in fact be due to cigarette smoking, a covariate highly correlated with alcohol consumption.4
With the flood of genetic association data published over the last few years, and the many others to come, it is the expectation that an increased number of the reported associations will be false findings or considered as such. As false finding is the common explanation for unexpected (irrelevant) but statistically significant associations, in most cases these findings are not followed-up. Nevertheless, associations appealing to common sense (association between marital status and cutaneous lymphoma) or as intriguing as they may appear (validated association between height and uterine fibroids) have been reported; these associations could be explained by mere LD, more precisely they might result from confounding by LD or GPD between loci influencing height and uterine fibroids.
Confounding by LD
To illustrate the notion of confounding by LD, let us consider the simple scenario of a Mendelian disease with complete or quasi-complete penetrance and a typed marker at gene locus 1 in LD with an untyped causal gene variant at gene locus 2. Let us further assume locus 1 affects a rare Mendelian form of obesity and locus 2 affects cancer but none of these gene functions is known. An epidemiological study that has collected measurements of potential confounders such as body mass index (BMI) may find this trait as an important predictor of cancer in the studied population, whereas the positive association may actually be explained by LD between the typed marker and the cancer-causing allele. Thus, LD can be a source of confounding as BMI predicts the cancer outcome in the absence of an apparent causal relationship. Fortunately, most of the genetic determinants of the common form of obesity identified and replicated so far explain only small fractions of the risk and therefore the impact of confounding by LD on association studies of common conditions may be limited but not negligible. By contrast, if the linked marker tags a gene with a sizeable effect, for example, a major gene, then confounding by LD can significantly affect the results of association studies, be they genetic or not. In our example above, if BMI is controlled for (that is, adjusted, stratified or restricted), the genetic effect of locus 2 on the cancer outcome cannot be accurately estimated and the observed association would typically be biased toward the null depending on the variance explained by locus 1 and the degree of LD between the two loci. In this case, the fitted model for the test of association may become overspecified, more precisely overadjusted because of adjustment for a covariate (BMI) influenced by a genetic locus (locus 1) in LD with the causal locus (locus 2). Most importantly, the covariate needs not to be on the causal pathway, bias toward the null can still be observed if gene locus 1 contributes significantly to the variance of the obesity trait in the study sample. Furthermore, because of the population-specific pattern of LD, confounding by LD is expected to vary across populations and thus could explain failures to replicate genetic association findings in comparable studies (those that controlled for the same confounders) in different populations.
Uterine fibroids and obesity
One of the illustrations of this phenomenon is the complex association between obesity and uterine leiomyomas (UL, also referred to as fibroids). Uterine fibroids are the most common tumors in women of reproductive age (about 75% of women will develop this condition by the time they reach menopause and 25% will have symptomatic fibroids). Cumulative exposure to estrogen is believed to be a major etiological factor5 and factors that may influence the hormonal milieu, such as obesity, are believed to be associated with risk.6 However, the clearly established risk factors are age (increasing risk with increasing premenopausal age), menopause (risk decreases with menopause) and African-American ethnicity (higher risk compared with that of non-Hispanic Whites). Epidemiological studies have shown mixed results with respect to the association between BMI and UL. Some studies reported positive or no associations, whereas other more adequately designed or larger studies reported an inverse J-shaped association,7, 8 with the association reaching a peak in the overweight category. The independent replication of this atypical association of BMI with UL in two cohort studies of UL7, 8 ruled out possible detection bias. In the absence of dose effect, plausible detection bias and homogeneity of the association across populations, a possible explanation for this atypical association is confounding by LD, that is, the presence of a major gene for obesity in the vicinity of a UL locus.
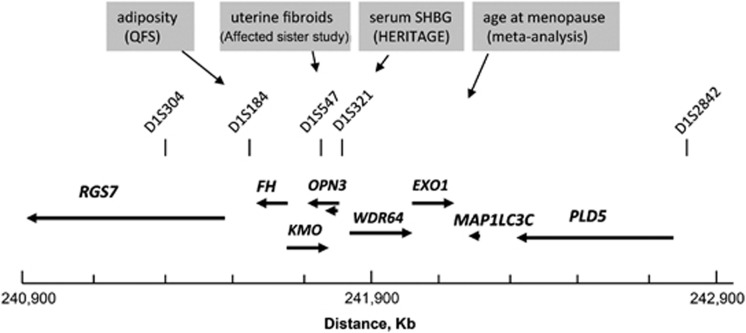
In a recent study, we have evaluated the role of chromosome 1q43 in the development of UL and we have identified two closely linked loci influencing differentially the risk of UL in European and African-American women.9 The peak of association with UL overlapped a 100 kb-long genomic region separating two head-to-tail genes encoding RGS7 (regulator of G-protein signaling 7) and FH (tricarboxylic acid cycle fumarate hydratase enzyme; Figure 1). Mutations in FH have previously been associated with two rare syndromic forms of UL, hereditary leiomyomatosis and renal cell cancer and multiple cutaneous and uterine leiomyomatosis.10, 11 The current paradigm associates FH to a tumor suppressor gene10, 11, 12 but several observations argue against it and models for an alternative 1q43 fibroid gene linked to FH and/or a pleiotropic FH have been postulated.9
Figure 1.
Colocalization of human traits of potential relevance to uterine fibroids on chromosome 1q43. Genomic map of a 2 Mb-long interval spanning the fumarate hydratase (FH) gene locus and showing the position of specific microsatellite markers (vertical bars) and gene loci linked or associated with diseases or traits of relevance to hormone-dependent tumors such as uterine leiomyomas. Quebec Family Study (QFS); sex hormone-binding globulin (SHBG); HERITAGE (HERITAGE Family Study). The placement of the genes in the gene map shown above the plot and the coordinates show below were derived the Human Genome assembly 19. Arrows indicate the orientation of the genes and are drawn proportionally to the size of the genes. RGS7 (regulator of G-protein 7); FH (fumarate hydratase); KMO (kynurenine 3-monooxygenase); OPN3 (opsin 3); WDR64 (WD repeat domain 64); EXO1 (exonuclease 1); MAP1LC3C (microtubule-associated protein 1 light chain 3 gamma); PLD5 (phospholipase D family, member 5).
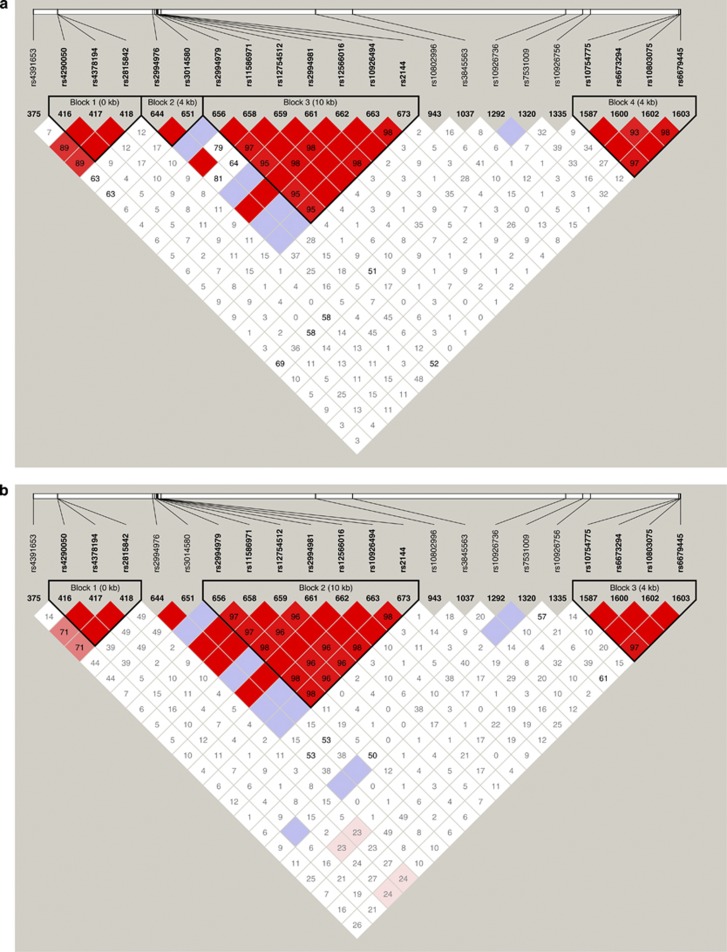
In an early study in the Quebec Family Study, we have mapped a quantitative trait locus for body fat to a chromosomal interval overlapping the FH locus.13 In the National Institute of Environmental Health Sciences Uterine Fibroid Study, we observed several signals for the association with BMI, with the most significant ones peaking in the RGS7-FH genomic interval in European Americans and in RGS7 and PLD5 (phospholipase D member 5) in African Americans (unpublished observation). However, it is still not clear which of FH, RGS7, PLD5 or another linked gene is the actual obesity gene because the association pattern across the FH-linked region varied by the UL affection status and race stratum. The emerging model is not in disagreement with our postulate of linked obesity and fibroid genes. Interestingly, the variance explained by the candidate 1q43 obesity gene is 20–30 times larger (r2=2–3%) than those reported for typical candidate obesity genes identified in meta-analyses of genome-wide association study.14 Nevertheless, the role of FH or the linked gene in non-syndromic UL is yet to be proven. As a proof-of-concept study for the phenomenon of confounding by LD, we analyzed the LD pattern among Chr.1q43 single nucleotide polymorphisms (SNPs) at risk for UL and/or obesity in the European group of the National Institute of Environmental Health Sciences Uterine Fibroid Study. Table 1 shows the list of 1q43 SNPs of interest, their relative position and the significance of their association with the UL and the BMI outcomes. To assess the independent effect of SNPs on each of the UL and BMI outcomes, we evaluated the association with UL in models with and without adjustment for BMI, and the association with BMI in models adjusted or not for the UL affection status. Confounding by LD is suspected if the LD pattern between SNPs associated with a given trait and SNPs associated with another trait is different among cases and controls. In our example, vanishing associations with UL at given ‘UL' SNPs after adjustment for BMI would be an indication for confounding due to LD with ‘obesity' SNPs. A direct demonstration of this phenomenon is suggested by the data in Figures 2a and b, which show a differential LD pattern at the tested ‘obesity' and ‘UL' SNPs in UL cases and controls, respectively. As can be seen, SNP375 (rs4391653) in RGS7 and SNP1320 (rs7531009) in PLD5, the two SNPs that remained significantly associated with BMI after controlling for the UL affection status, show different levels of LD with the ‘UL' SNPs in cases and controls. Specifically, the SNPs at which the association was lost after adjustment for BMI (RGS7 SNPs 416–418 and RGS-FH intergenic SNPs 644 and 651) exhibit opposite levels of LD with the two ‘obesity' SNPs 375 and 1320 in the UL cases compared with the controls.
Table 1. Association of 1q43 SNPs with uterine fibroids and BMI in European Americans enrolled in the NIEHS study.
|
Uterine fibroids (UL), Pa |
BMI, Pb |
|||||||
|---|---|---|---|---|---|---|---|---|
| Obs | SNP | Inter-SNP distance, base pair | Gene | Function | BMI adjusted | BMI unadjusted | UL adjusted | UL unadjusted |
| 375 | rs4391653 | RGS7 | Intronic | 0.0063 | 0.0272 | 0.0245 | ||
| 416 | rs4290050 | 57 222 | RGS7 | Intronic | 0.0078 | |||
| 417 | rs4378194 | 19 | RGS7 | Intronic | 0.0153 | |||
| 418 | rs2815842 | 229 | RGS7 | Intronic | 0.0143 | |||
| 644 | rs2994976 | 225 065 | RGS7-FH | Intergenic | 0.0482 | |||
| 651 | rs3014580 | 4859 | RGS7-FH | Intergenic | 0.0457 | |||
| 656 | rs2994979 | 4046 | Downstream FH | Intronic | 0.0050 | 0.0800 | ||
| 658 | rs11586971 | 1123 | Downstream FH | Intronic | 0.0050 | |||
| 659 | rs12754512 | 51 | Downstream FH | Intronic | 0.0050 | |||
| 661 | rs2994981 | 1967 | downstream FH | Intronic | 0.0045 | |||
| 662 | rs12566016 | 460 | downstream FH | Intronic | 0.0045 | |||
| 663 | rs10926494 | 204 | downstream FH | Intronic | 0.0017 | |||
| 673 | rs2144 | 7176 | Downstream FH | Intronic | 0.0014 | |||
| 943 | rs10802996 | 365 915 | EXO1 | 5′-UTR | 0.0075 | |||
| 948 | rs4149864 | 58 417 | EXO1-MAP1LC3C | Intergenic | 0.0724 | 0.0274 | ||
| 987 | rs4150028 | 29 371 | EXO1-MAP1LC3C | Intergenic | 0.0193 | |||
| 1037 | rs3845563 | 545 213 | PLD5 | Intronic | 0.0016 | |||
| 1292 | rs10926736 | 10 854 | PLD5 | Intronic | 0.0019 | |||
| 1320 | rs7531009 | 7558 | PLD5 | Intronic | 0.0064 | 0.0098 | 0.0099 | |
| 1335 | rs10926756 | 208 978 | Far upstream PLD5 | Intergenic | 0.0015 | |||
| 1340 | rs1039534 | 1531 | Far upstream PLD5 | Intergenic | 0.0047 | |||
| 1587 | rs10754775 | 160 | Far upstream PLD5 | Intergenic | 0.0047 | |||
| 1600 | rs6673294 | 1757 | Far upstream PLD5 | Intergenic | 0.0043 | |||
| 1602 | rs10803075 | 493 | Far upstream PLD5 | Intergenic | 0.0053 | |||
| 1603 | rs6679445 | 494 | Far upstream PLD5 | Intergenic | 0.0014 | |||
Abbreviations: BMI, body mass index; FH, fumarate hydratase; PDL5, phospholipase D member 5; SNP, single nucleotide polymorphism; UL, uterine leiomyoma.
Logistic regression models adjusted for covariates (age, age at menarche, parity and physical activity).
General linear models adjusted for the effects of age and physical activity. Note that only SNPs that showed significant levels of association with either the uterine fibroid or the BMI outcome or with both outcomes are shown.
Figure 2.
Pattern of linkage disequilibrium (LD) between single nucleotide polymorphisms (SNPs) at risk for uterine fibroids and/or obesity in a case (panel a) and control (panel b) study of uterine fibroids in the National Institute of Environmental Health Sciences Uterine Fibroid Study. Note that in contrast to the ‘uterine leiomyoma (UL)' SNPs in the region downstream FH (SNP 656–673), which show low levels of or no LD with the two ‘obesity' SNPs, the association with SNP 1037, 1292 and 1335 is lost after controlling for body mass index (BMI). A full color version of this figure is available at the Journal of Human Genetics journal online.
Although the current data support the presence of colocalized susceptibility loci for obesity13, 15, 16 and reproduction-related traits,9, 10, 11, 17, 18 it remains to be seen whether the present proof-of-concept study will be strengthen or weakened following gene identification and mutational analyses. In our example, the use of tag SNPs as proxies for the actual causal variants and BMI as proxy for body fat to assess genetic confounding by LD may mask the true impact of this phenomenon. Consequently, confirmation and validation of this hypothesis in a genomic context with known and validated genotype–phenotype relationships should be an important undertaking.
In a meta-analysis study of age at natural menopause in populations of European descent, EXO1, a DNA repair gene that maps close to FH was highlighted as one of the 13 loci influencing this outcome.18 Mapping of age at natural menopause, one of the established risk factors for UL, to a region implicated in UL and obesity makes the potential of confounding by LD more plausible. Thus, correlated traits may provide clues to the location of disease genes but can also confound the association. Actually, the UL example is not a simple illustration of genetic confounding by LD because UL is a hormonally dependent cancer believed to be influenced by steroid bioavailability, a level of which is in turn influenced by the body fat through downregulation of circulating sex hormone-binding globulin.19 Coincidentally, linkage for the circulating level of sex hormone-binding globulin in the HERITAGE study peaked at D1S321, a marker within 150–200 kb distance from FH.20 Thus, further complications arise as the linked candidate obesity gene may affect the association with fibroids in several ways: through the causal pathway by decreasing the serum level of sex hormone-binding globulin and consequent increase in bioavailability of free steroids, confounding by LD or through combination of both.
LD and selection
On the other hand, confounding by LD can be useful for association studies (gain of statistical power in bivariate models) because genes that have similar and/or coordinated functions tend to be clustered in the genome of eukaryotes.21 In the obesity and UL example, it is tempting to think that the atypical association between obesity and fibroids may actually reflect evolutionary selective constraints on ‘thrifty' obesity and reproduction-associated genes that have evolved different mutation patterns in the history of human populations. The selective advantages of the overweight-to-obese traits have amply been discussed in the past;22 here I propose a thrifty phenotype model for the high incidence of UL in women. Reproductive suppression as an adaptive response to low-energy availability23 would be an attractive evolutionary model for UL. With reproduction and metabolism (fat storage) representing the most selectively constrained traits and functions, occurrence and conservation of LD between their genetic determinants in populations are alike. In this line of thought, a large-scale study (CARe—the National Heart, Lung, and Blood Institute Candidate Gene Association Resource) of ultraconserved polymorphisms in the human genome, which are believed to affect reproductive (age at natural menopause, number of children, age at first child and age at last child) and overall (longevity, BMI and height) fitness,24, 25 has shown an excess of associations with BMI.26 Interestingly, the most strongly associated SNP, rs10818872, occurred in DENND1A, a locus previously shown to be associated with polycystic ovary syndrome and detected by a SNP in high LD (r2=0.826) with rs10818872.27 Independently, however, whether the atypical association of BMI and UL is real or artifactual, and whether it is related to the candidate 1q43 region or not, confounding by LD should be real and a valid notion potentially explaining the growing reports on shared genetic polymorphisms between obesity- and reproduction-related traits28, 29, 30, 31, 32, 33 including those implicating chromosome 1q43.9, 18 Similarly, follow-up of genome-wide linkage and admixture signals for UL in the NIEHS cohort pointed to several known obesity-related genes (Aissani et al., unpublished observation).
Confounding by GPD
The occurrence of GPD may reflect demographic patterns or can be the result of evolutionary processes that favored interactions between genes (epistasis) contributing to the expression of specific traits in populations under specific environments. Similarly, GPD can confound association tests and may account for a number of the claimed associations with common diseases (for example, 349 hits with genome-wide distribution were found in a query of the Map viewer database—NCBI—with the keyword ‘obesity'). Studies exploring the genetic basis of correlated traits and clinical phenotypes in relationship to coordinated epistatic gene expression are still in their infancy and new integrated approaches to tackle this complexity are needed. The existence of trans-regulation of expression phenotypes at the genome level34 further suggests that GPD may underlie the correlation among human traits through coordinated gene expressions In this context, the emerging field of phenomics and its combination with genome-wide association study in the so-called phenome-wide association studies is an important advance in the design of the next generation of genetic association studies of common diseases.35 Furthermore, the development of resources such as Population Architecture using Genomics and Epidemiology—National Human Genome Research Institute to characterize well-replicated genome-wide association study variants in relationship to many traits and disease phenotypes36 will provide opportunities to test new hypotheses on the genetic basis of correlated traits and comorbidities.
Confounding by pleiotropy
The other source of genetic confounding is obviously pleiotropy (one gene or one mutation affecting more than one trait or phenotype), a concept known for exactly a century37 but the interference of this phenomenon in genetic association studies has not been fully investigated. Empirical data essentially from model organisms indicated that pleiotropy is a common phenomenon.38 At least in yeast, pleiotropy is believed to be mostly of type 2, that is, a single molecular function resulting in multiple effects (as opposed to type 1 pleiotropy, which is one gene, multiple functions).39 As outlined in the UL example, it is not yet clear whether the UL gene on 1q43 is pleiotropic (an obesity gene affecting UL through change in hormonal milieu), a hypothesis supported by the linkage of the FH region to the level of circulating sex hormone-binding globulin in the HERITAGE study, or a UL gene confounded by a linked obesity gene. In either scenario, FH remains a strong candidate pleiotropic gene. Indeed, a model has been proposed whereby a single inactivating mutation can affect distinct functions encoded by FH,9 with cytosolic FH possibly acting in DNA repair activity40 and mitochondrial FH in metabolism. Models that analyze the effects of genetic variation on the combined outcomes (bivariate or multivariate analyses) theoretically should increase the power to capture the underlying pleiotropic loci.
Confounding by cytonuclear LD
Confounding by yet another type of LD, cytonuclear LD (for LD between DNA variants present in organelles and in the nucleus), is more challenging and remains largely unexplored. A priori, any positive associations with nuclear variants may actually be proxies for true causal mitochondrial variants and vice versa. Confounding by cytonuclear LD can be more subtle, given that as many as 966 or more nuclear-encoded factors are involved in the maintenance and function of the mitochondrion. Thus, mitochondrial diseases can have diverse modes of inheritance (maternal, Mendelian and a combination of the two) with variable expression of their phenotypes owing to the stochastic and quantitative nature of inheritance of heteroplasmic mutations.41 As a result, current knowledge and methodological approaches to evaluate the joint effects of co-inherited mitochondrial and nuclear variants on human diseases are limited. For diseases such as cancer, infection and immune inflammatory diseases, the interaction effects of specific nuclear and apoptosis-related mitochondrial variants are likely to be important determinants of disease risk or progression. One possible way to control for the effects of mitochondrial variants in genome-wide association study is to type diagnostic mitochondrial polymorphisms to allow for the mitochondrial haplogroup membership to be inferred and used as a covariate.42
The interplay between the nuclear and the mitochondrial genomes is likely to be central in the regulation of energy homeostasis and perturbations in the coordinated expression of the underlying nuclear and mitochondrial genes may lead to poor cellular performances and diseases. Conceivably, recent population admixtures may increase the risk for certain conditions that arise from selectively constrained human traits such as obesity, reproduction and resistance to infection, hence reaching genetic fitness in the parental populations of recently admixed populations.
Conclusion
The correlations among traits and consequently among clinical phenotypes undoubtedly have clouded the interpretation of many genetic association data. Beyond the initial quest to highlight aspects of LD that may affect the interpretation of association studies, I extended the reasoning to convey new thoughts on the possible existence of thrifty phenotypes to account for the high incidence of uterine fibroids and to some extent of reproductive cancer in general in human populations. Confounding by LD or GPD is a novel concept that may in part explain failures to replicate findings in genetic associations that incorporate covariates differentially correlated in different populations. Finally, while perplexing but legitimate interrogations have been raised in this paper on the meaning of genome–phenome associations in the context of dynamic genomes, coordinated gene expressions and correlated traits and diseases, a framework of thought for genetically and evolutionary determined ‘thrifty correlated phenotypes' is suggested to account for the high incidence of uterine fibroids in the contemporary obesogenic environment.
Acknowledgments
I am indebted to Dr Donna Baird for sharing the data and the biospecimens collected for the NIEHS Uterine Fibroid Study and would like to thank the participants to this study. This study was funded by NIH Grant R01-HD064398 to BA from the National Institute of Child and Health Development (NIH-NICHD) and National Institute for Environmental and Health Science (NIH-NIEHS).
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Wang X., Elston R. C., Zhu X. The meaning of interaction. Hum. Hered. 2010;70:269–277. doi: 10.1159/000321967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asmussen M. A., Arnold J., Avise J. C. Definition and properties of disequilibrium statistics for associations between nuclear and cytoplasmic genotypes. Genetics. 1987;115:755–768. doi: 10.1093/genetics/115.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward M.Epidemiology: Study Design and Data Analysis, 2nd ed2nd edn (Chapmanand Hall/CRC: Boca Raton, FL, USA; 2004 [Google Scholar]
- Dales L. G., Ury H. K. An improper use of statistical significance testing in studying covariables. Int. J. Epidemiol. 1978;7:373–375. doi: 10.1093/ije/7.4.373. [DOI] [PubMed] [Google Scholar]
- Andersen J. Growth factors and cytokines in uterine leiomyomas. Semin. Reprod. Endocrinol. 1996;14:269–282. doi: 10.1055/s-2007-1016336. [DOI] [PubMed] [Google Scholar]
- Schwartz S. M., Marshall L. M., Baird D. D. Epidemiologic contributions to understanding the etiology of uterine leiomyomata. Environ. Health Perspect. 2000;108 (Suppl 5:821–827. doi: 10.1289/ehp.00108s5821. [DOI] [PubMed] [Google Scholar]
- Baird D. D., Dunson D. B., Hill M. C., Cousins D., Schectman J. M. Association of physical activity with development of uterine leiomyoma. Am. J. Epidemiol. 2007;165:157–163. doi: 10.1093/aje/kwj363. [DOI] [PubMed] [Google Scholar]
- Wise L. A., Palmer J. R., Spiegelman D., Harlow B. L., Stewart E. A., Adams-Campbell L. L., et al. Influence of body size and body fat distribution on risk of uterine leiomyomata in U.S. black women. Epidemiology. 2005;16:346–354. doi: 10.1097/01.ede.0000158742.11877.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aissani B., Wiener H., Zhang K. Multiple hits for the association of uterine fibroids on human chromosome 1q43. PLoS One. 2013;8:e58399. doi: 10.1371/journal.pone.0058399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam N. A., Bevan S., Churchman M., Barclay E., Barker K., Jaeger E. E., et al. Localization of a gene (MCUL1) for multiple cutaneous leiomyomata and uterine fibroids to chromosome 1q42.3-q43. Am. J. Hum. Genet. 2001;68:1264–1269. doi: 10.1086/320124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomlinson I. P., Alam N. A., Rowan A. J., Barclay E., Jaeger E. E., Kelsell D., et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat. Genet. 2002;30:406–410. doi: 10.1038/ng849. [DOI] [PubMed] [Google Scholar]
- Lehtonen R., Kiuru M., Vanharanta S., Sjöberg J., Aaltonen L. M., Aittomäki K., et al. Biallelic inactivation of fumarate hydratase (FH) occurs in nonsyndromic uterine leiomyomas but is rare in other tumors. Am. J. Pathol. 2004;164:17–22. doi: 10.1016/S0002-9440(10)63091-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aissani B., Perusse L., Lapointe G., Chagnon Y. C., Bouchard L., Walts B., et al. A quantitative trait locus for body fat on chromosome 1q43 in French Canadians: linkage and association studies. Obesity. 2006;14:1605–1615. doi: 10.1038/oby.2006.185. [DOI] [PubMed] [Google Scholar]
- Kilpelainen T. O., Zillikens M. C., Stancakova A., Finucane F. M., Ried J. S., Langenberg C., et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat. Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosse Y., Chagnon Y. C., Despres J. P., Rice T., Rao D. C., Bouchard C., et al. Genome-wide linkage scan reveals multiple susceptibility loci influencing lipid and lipoprotein levels in the Quebec Family Study. J. Lipid Res. 2004;45:419–426. doi: 10.1194/jlr.M300401-JLR200. [DOI] [PubMed] [Google Scholar]
- Dong C., Beecham A., Slifer S., Wang L., McClendon M. S., Blanton S. H., et al. Genome-wide linkage and peak-wide association study of obesity-related quantitative traits in Caribbean Hispanics. Hum. Genet. 2011;129:209–219. doi: 10.1007/s00439-010-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross K. L., Panhuysen C. I., Kleinman M. S., Goldhammer H., Jones E. S., Nassery N., et al. Involvement of fumarate hydratase in nonsyndromic uterine leiomyomas: genetic linkage analysis and FISH studies. Genes Chromosomes Cancer. 2004;41:183–190. doi: 10.1002/gcc.20079. [DOI] [PubMed] [Google Scholar]
- Stolk L., Perry J. R., Chasman D. I., He C., Mangino M., Sulem P., et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat. Genet. 2012;44:260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorgan J. F., Reichman M. E., Judd J. T., Brown C., Longcope C., Schatzkin A., et al. Relation of energy, fat, and fiber intakes to plasma concentrations of estrogens and androgens in premenopausal women. Am. J. Clin. Nutr. 1996;64:25–31. doi: 10.1093/ajcn/64.1.25. [DOI] [PubMed] [Google Scholar]
- Ukkola O., Rankinen T., Gagnon J., Leon A. S., Skinner J. S., Wilmore J. H., et al. A genome-wide linkage scan for steroids and SHBG levels in black and white families: the HERITAGE Family Study. J. Clin. Endocrinol. Metab. 2002;87:3708–3720. doi: 10.1210/jcem.87.8.8725. [DOI] [PubMed] [Google Scholar]
- Hurst L. D., Pal C., Lercher M. J. The evolutionary dynamics of eukaryotic gene order. Nat. Rev. Genet. 2004;5:299–310. doi: 10.1038/nrg1319. [DOI] [PubMed] [Google Scholar]
- Neel J. V. Diabetes mellitus: a ‘thrifty' genotype rendered detrimental by ‘progress'. Am. J. Hum. Genet. 1962;14:353–362. [PMC free article] [PubMed] [Google Scholar]
- Jasienska G. Energy metabolism and the evolution of reproductive suppression in the human female. Acta Biotheor. 2003;51:1–18. doi: 10.1023/a:1023035321162. [DOI] [PubMed] [Google Scholar]
- Derti A., Roth F. P., Church G. M., Wu C. T. Mammalian ultraconserved elements are strongly depleted among segmental duplications and copy number variants. Nat. Genet. 2006;38:1216–1220. doi: 10.1038/ng1888. [DOI] [PubMed] [Google Scholar]
- Ahituv N., Zhu Y., Visel A., Holt A., Afzal V., Pennacchio L. A., et al. Deletion of ultraconserved elements yields viable mice. PLoS Biol. 2007;5:e234. doi: 10.1371/journal.pbio.0050234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang C. W., Liu C. T., Lettre G., Lange L. A., Jorgensen N. W., Keating B. J., et al. Ultraconserved elements in the human genome: association and transmission analyses of highly constrained single-nucleotide polymorphisms. Genetics. 2012;192:253–266. doi: 10.1534/genetics.112.141945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. J., Zhao H., He L., Shi Y., Qin Y., Shi Y., et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat. Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- Elks C. E., Perry J. R., Sulem P., Chasman D. I., Franceschini N., He C., et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat. Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Rhodes L., Demerath E. W., Cousminer D. L., Tao R., Dreyfus J. G., Esko T., et al. Association of adiposity genetic variants with menarche timing in 92,105 women of European descent. Am. J. Epidemiol. 2013;178:451–460. doi: 10.1093/aje/kws473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed M. L., Ong K. K., Dunger D. B. Childhood obesity and the timing of puberty. Trends Endocrinol. Metab. 2009;20:237–242. doi: 10.1016/j.tem.2009.02.004. [DOI] [PubMed] [Google Scholar]
- Frayling T. M., Ong K. Piecing together the FTO jigsaw. Genome Biol. 2011;12:104. doi: 10.1186/gb-2011-12-2-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hoed M., Ekelund U., Brage S., Grontved A., Zhao J. H., Sharp S. J., et al. Genetic susceptibility to obesity and related traits in childhood and adolescence: influence of loci identified by genome-wide association studies. Diabetes. 2010;59:2980–2988. doi: 10.2337/db10-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Bradfield J. P., Li M., Wang K., Zhang H., Kim C. E., et al. The role of obesity-associated loci identified in genome-wide association studies in the determination of pediatric BMI. Obesity. 2009;17:2254–2257. doi: 10.1038/oby.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goring H. H., Curran J. E., Johnson M. P., Dyer T. D., Charlesworth J., Cole S. A., et al. Discovery of expression QTLs using large-scale transcriptional profiling in human lymphocytes. Nat. Genet. 2007;39:1208–1216. doi: 10.1038/ng2119. [DOI] [PubMed] [Google Scholar]
- Denny J. C., Ritchie M. D., Basford M. A., Pulley J. M., Bastarache L., Brown-Gentry K., et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matise T. C., Ambite J. L., Buyske S., Carlson C. S., Cole S. A., Crawford D. C., et al. The Next PAGE in understanding complex traits: design for the analysis of Population Architecture Using Genetics and Epidemiology (PAGE) Study. Am. J. Epidemiol. 2011;174:849–859. doi: 10.1093/aje/kwr160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns F. W. One hundred years of pleiotropy: a retrospective. Genetics. 2010;186:767–773. doi: 10.1534/genetics.110.122549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner G. P., Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat. Rev. Genet. 2011;12:204–213. doi: 10.1038/nrg2949. [DOI] [PubMed] [Google Scholar]
- He X., Zhang J. Toward a molecular understanding of pleiotropy. Genetics. 2006;173:1885–1891. doi: 10.1534/genetics.106.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O., Yogev O., Singer E., Shaulian E., Golberg M., Fox T. D., et al. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010;8:e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace D. C. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- Johnson D. C., Shrestha S., Wiener H. W., Makowsky R., Kurundkar A., Wilson C. M., et al. Mitochondrial DNA diversity in the African American population. Mitochondrial DNA. 2013;24:613–615. doi: 10.3109/19401736.2013.840591. [DOI] [PMC free article] [PubMed] [Google Scholar]