Figure 1.
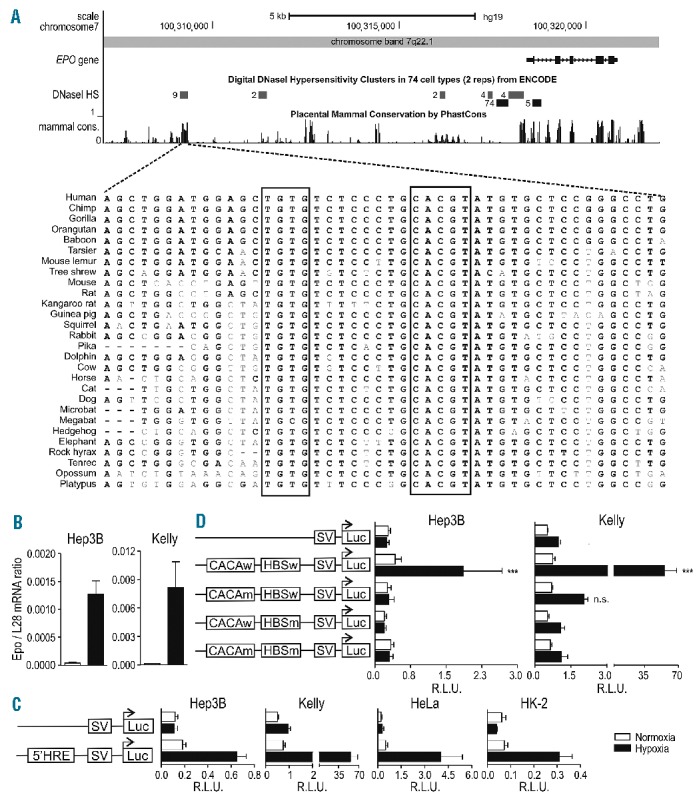
Location of a functional distal HRE in the Epo 5′ regulatory region. (A) UCSC Genome Browser output (hg19) of the Epo genomic and 5′ upstream region. Shown are the ENCODE DSS clusters and mammalian PhastCons conservation tracks with a closer view of the region in 28 vertebrates extracted using the 46-MULTIZ whole-genome multiple alignment algorithm. (B) Epo mRNA levels in human Hep3B and Kelly cells were measured by RT-qPCR and normalized to ribosomal protein L28 mRNA levels. (C) Both cell types (3×105 cells) were co-transfected with the indicated SV40-driven firefly luciferase reporter gene plasmids (500 ng) and a Renilla luciferase control plasmid (5 ng, Promega) in a 6-well format by polyethylenimine (PEI). 24 h post-transfection, cells were incubated for another 24 h under normoxic or hypoxic (0.2% O2) conditions. Luciferase activities of triplicate wells were determined using the Dual Luciferase Reporter Assay System according to the manufacturer’s protocol (Promega). All results are displayed as ratios of firefly to Renilla relative light units (R.L.U.). (D) Cells were co-transfected with pGL3p Epo 5′-HRE wild-type (w), pGL3p Epo HBS mutated (m) (ACGTG to AAAAG), pGL3p Epo CACA mutated (CACA to AAAA) or pGL3p Epo HBSm, CACAm together with a Renilla luciferase control plasmid, and reporter gene experiments performed as in D. (B–D) All data are expressed as mean ± SEM of 3 independent experiments and statistical analyses were performed with one-way ANOVA and Tukey correction for multiple comparisons (*P<0.05; **P<0.01; ***P<0.001).
