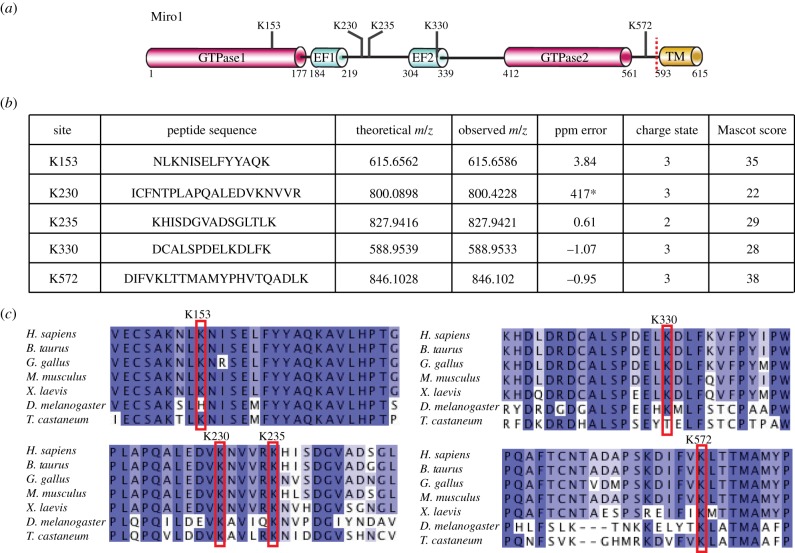
Figure 3.
Parkin ubiquitylates Miro1 at multiple sites in a PINK1-dependent manner. (a) A schematic of Miro1 domain architecture showing the identified ubiquitylation sites and truncation site (red dotted line) used in this paper. (b) Identification of Lys153, Lys230, Lys235, Lys330 and Lys572 ubiquitylation sites on Miro1. Ubiquitylation assays using wild-type (WT) and kinase-inactive (KI) PINK1 (D359A) in combination with WT Parkin and the substrate Miro1 were undertaken as described in §3. The SDS–PAGE bands were subjected to in-gel tryptic digestion and analysis by LTQ-Orbitrap mass spectrometry. Ubiquitin isopeptides were identified by Mascot (www.matrixscience.com), and spectra were manually validated to ensure peptide fragmentation gave good sequence coverage (*417 ppm error equates to a −1.81 ppm error around the C13 isotope). (c) Sequence alignment of residues around Lys153, Lys230 and Lys235 (left), Lys330 and Lys572 (right), respectively, in human Miro1 and a variety of lower organisms, showing high degree of conservation.