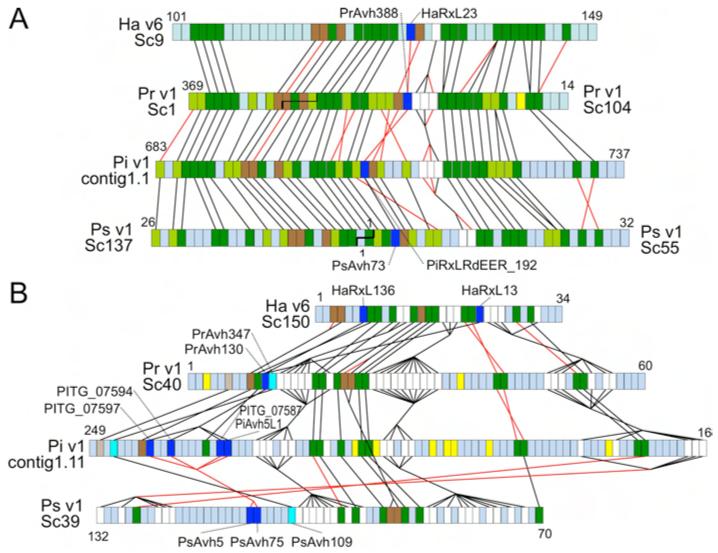
Fig. 2.
Synteny of conserved RXLR effectors. (A) Region around HaRxL23, spanning scaffold_9:467737- 739923 (v6) and supercontig16:325445- 40488 (v8.3.2) (B) Region around HaRxL13 and HaRxL136, spanning scaffold_150: 3503-183330 (v6) and supercontig35:456072-268293 (v8.3.2). Colored boxes show order of gene models. Non-coding DNA is not represented. Dark green, orthologs; light green, orthologs found only in Phytophthora; dark brown, syntenic paralogs; light brown, syntenic orthologs found only in Phytophthora; white, syntenic gene families; dark blue, syntenic conserved RXLR effectors; cyan, syntenic RXLR effectors conserved only in Phytophthora; yellow, RXLR effectors not syntenic or conserved; blue-gray, other genes not conserved or syntenic. Black lines join syntenic genes with the same orientation; red lines join genes with reversed orientations. Staggered black lines in (A) show scaffold joins predicted from the synteny analysis. HaRxL23, HaRxL13 and HaRxL136 have 47%, 38% and 40% amino acid identity, respectively, with their most similar Phytophthora ortholog, within the normally hypervariable C terminus.