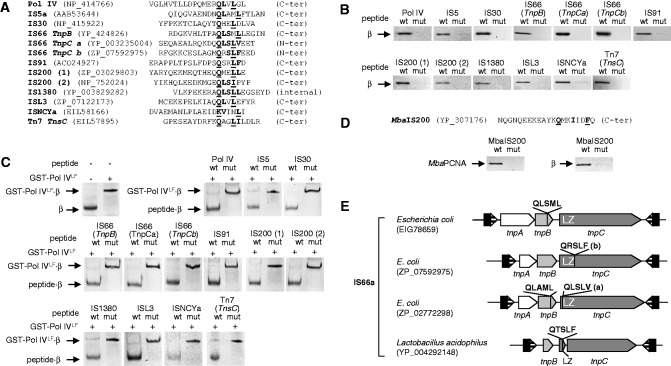
Fig. 3.—
Interaction between transposases and the β sliding clamp. (A) List of peptides used in the binding assays, aligned at the β-binding motif. The Pol IV peptide was used as a positive control. Peptides derived from transposases found in Escherichia coli chromosomes. Residues corresponding to the consensus β motif are in bold type and those mutated to alanine, underlined. Two peptides (a and b) were designed for different regions of the TnpC protein of IS66a (see E). Two homologous peptides (1 and 2) were designed corresponding to variants of IS200 transposase. NCBI accession numbers for protein sequences are shown. (B) Peptides were coupled to streptavidin-coated paramagnetic beads and used to retain purified Alexa 350-labeled E. coli β. In each panel, the native (wt) and the mutated (mut) peptides were used. (C) Fluorescently labeled β whose mobility was retarded in a native gel by interaction with GST-Pol IVLF was challenged with an excess of transposase-derived peptides, as indicated (See Materials and Methods). (D) N-biotinylated peptides derived from the C-terminus of Methanosarcina barkeri IS200 were bound to streptavidin-coated magnetic beads as in B and used to probe interaction with M. barkeri PCNA (left panel) or E. coli β (right panel). The PCNA consensus motif (bold) and the residues mutated in the mutated peptide (underlined) are marked. (E) Structure of IS66 and diversity in sequence and location of β-binding motifs. The motif can be present in TnpB, TnpC, or in both, and in TnpC, it can be located upstream or downstream of the leuzine zipper domain (LZ). In some Bacilli (Firmicutes), the LZ domain is an independent open reading frame and presents the β-binding motif at its C-terminus (see supplementary table S5, Supplementary Material online).