Figure 9.
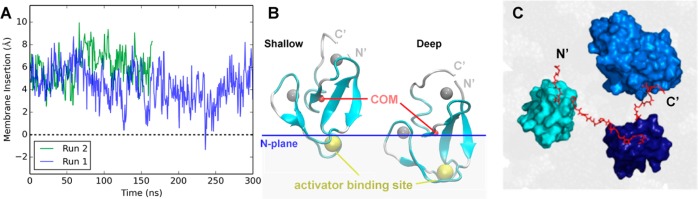
All-atom MD simulations of the isolated C1A domain and the C1A–C1B–C2 construct bound to a PC/PS bilayer. (A) Membrane insertion [measured every 0.54 ns as the distance from the center of mass (COM) of the protein to the bilayer PC average N plane] over time observed in two production runs. (B) Cartoon representation of the C1A domain (cyan) showing the zinc ions (silver spheres), the protein center of mass (COM, red sphere), and the bilayer N plane as a blue line. At the left is a snapshot of the shallow state (8.7 Å) at 59 ns of run 2 and at the right a snapshot of the C1A deep state (0.3 Å) at 245 ns of run 1. A detailed conformational analysis shows that three lysines (K45, K62, and K76) and two arginines (R42 and R77) are the major residues involved in the electrostatic interactions between the C1A domain and the PC/PS headgroups in the shallow state. Four of these residues (R42, K62, K76, and R77) were observed to form a charged “semicircle” around the middle of the C1A domain to stabilize the deep insertion state. It is noteworthy that in the deep state, a stable hydrogen bond is formed between K45 and the backbone oxygen atom of F43 to stabilize the buried charge, while cation−π interactions between side chains of K45 and F43 or F44 may also contribute to the stability of K45 in the deep state. In addition, >60% of the C1A conformers in the deep state were associated with PS, while no shallow state conformers were observed to bind PS. (C) View down the membrane normal toward the bilayer surface of a C1A–C1B–C2 construct (surface representation) on the bilayer (gray) after a 10 ns all-atom MD simulation. The simulation was initiated with the preferred membrane docking geometry of each individual domain (Figure 9B and ref (36)), and with the interdomain linkers (red sticks, H and side-chain atoms hidden) extended with no steric clash. The resulting model was stable in the membrane during the entire simulation. N′ and C′ represent the truncated N- and C-terminal ends of the fragments employed in both experimental studies (Figure 2) and MD simulations.
