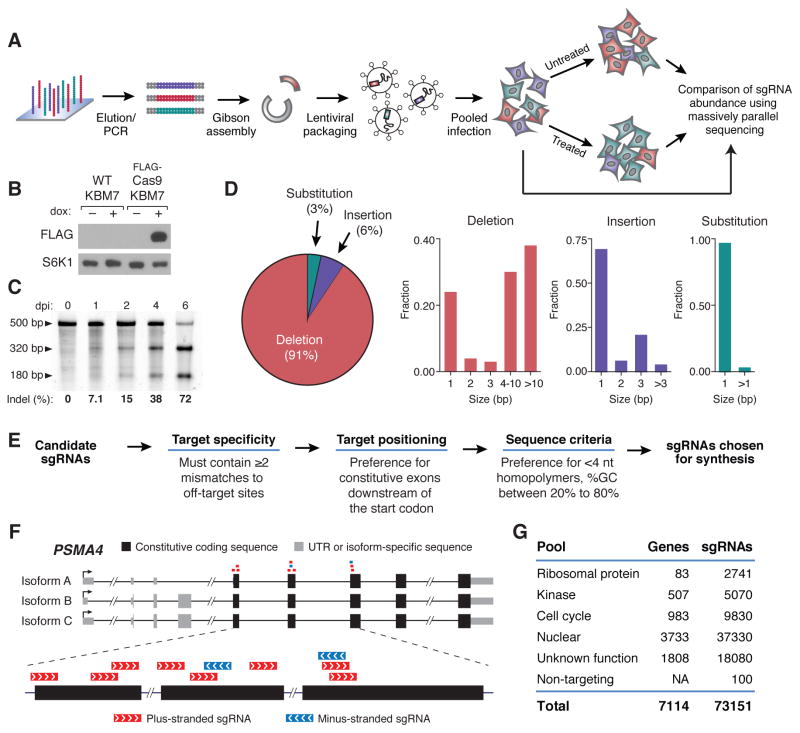
Fig. 1. A pooled approach for genetic screening in mammalian cells using a lentiviral CRISPR/Cas9 system.
(A) Outline of sgRNA library construction and genetic screening strategy (B) Immunoblot analysis of wild-type KBM7 cells and KBM7 cells transduced with a doxycycline inducible FLAG-Cas9 construct upon doxycycline induction. S6K1 was used as a loading control. (C) Sufficiency of single copy sgRNAs to induce genomic cleavage. Cas9-expressing KBM7 cells were transduced with AAVS1-targeting sgRNA lentivirus at low MOI. The SURVEYOR mutation detection assay was performed on cells at the indicated days post-infection (dpi). Briefly, mutations resulting from cleavage of the AAVS1 locus were detected through PCR amplification of a 500-bp amplicon flanking the target sequence, re-annealing of the PCR product and selective digestion of mismatched heteroduplex fragments. (D) Characterization of mutations induced by CRISPR/Cas9 as analyzed by high-throughput sequencing. (E) sgRNA library design pipeline. (F) Example of sgRNAs designed for PSMA4. sgRNAs targeting constitutive exonic coding sequences nearest to the start codon were chosen for construction. (G) Composition of genome-scale sgRNA library.