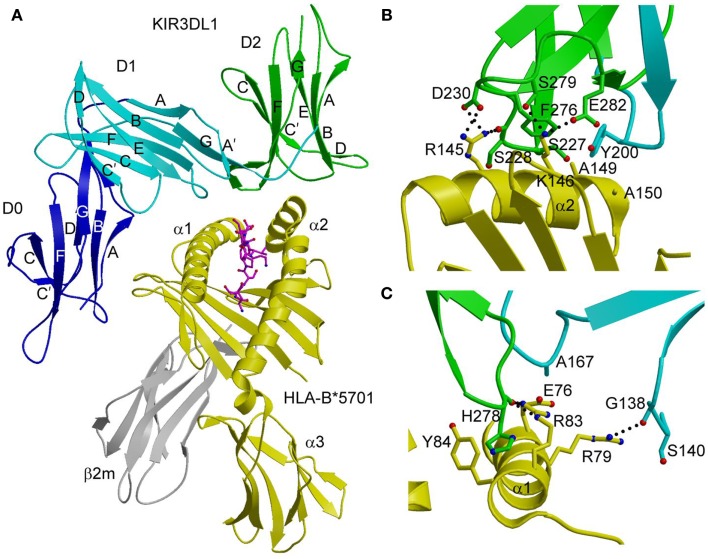
Figure 2.
Structure of the KIR3DL1–HLA-B*5701 complex. (A) Ribbon diagram of KIR3DL1 bound to HLA-B*5701 (3VH8). The orientation of the MHC-I ligand is similar to that of HLA-Cw3 in the KIR2DL2–HLA-Cw3 complex (Figure 1B). The HLA-B*5701 heavy chain is yellow; β2m is gray; the peptide is magenta. The KIR3DL1 D0 domain is dark blue; D1 is cyan; D2 is green. The secondary structural elements of KIR3DL1 are labeled. (B) Contacts between KIR3DL1 and the HLA-B*5701 α2 helix. The D2 domain mainly interacts with HLA-B*5701 residues 142–151, which display limited polymorphism among HLA-B alleles. At the center of the D2–HLA-B*5701 interface, KIR3DL1 residues Tyr200 and Phe276 form an aromatic cluster that converges on the α2 helix. (C) Contacts between KIR3DL1 and the HLA-B*5701 α1 helix. KIR3DL1 recognizes HLA allotypes that contain the Bw4 epitope-defining residues 77–83 on the α1 helix, which likely accounts for the allelic specificity of KIR3DLs.