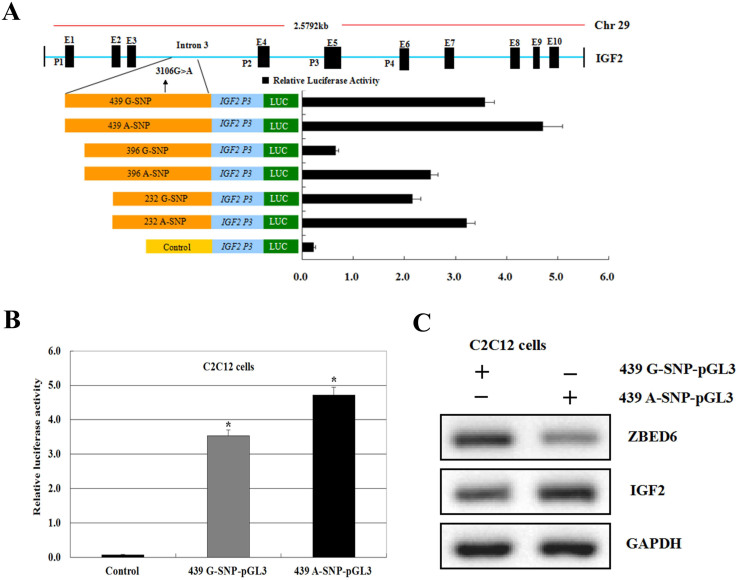
Figure 5. Recombinant vectors used to assay the sequence activity of IGF2 gene intron 3.
(A) Luciferase activity of IGF2 (Black bars) intron 3 sequence 3106G>A mutant constructs in C2C12 cells. The location and size of each fragment is indicated to the left of each bar relative to the intron 3 start codon. IGF2 promoter activity modulated by genetic polymorphisms (intron 3–3106G>A). Luciferase activity were transfected with recombinant plasmids containing the intron 3–3106 G-SNP and −3106 A-SNP and the bovine IGF2 P3 promoter in cell lines. Results from pGL3-Basic plasmid are given as a negative control. (B) Comparison of luciferase activity levels of IGF2 in C2C12 cell lines between transfected with pcDNA3.1+ (control), pcDNA3.1+-ZBED6. The blank expression vector (pcDNA3.1+ and pGL3-Basic) was used to maintain equivalent amounts of DNA. Gray solid bar show luciferase activities from cell lines transfected with the wild type haplotype IGF2 439G-SNP-pGL3 construct containing the wild-type allele. Black solid bar represent luciferase activities from cell lines transfected with the mutant haplotype 439A-SNP-pGL3 construct containing the mutant allele. Data are mean ± SEM of normalized luciferase activity. (Values represent the mean ± SEM of three duplications.) *P < 0.05. (C) Crude proteins were extracted from the treated cells and then subjected to Western blot with antibodies against ZBED6 and IGF2, GAPDH was used as a control for equal protein loading. Data are representative of at least three independent experiments performed in duplicate (n = 3 or n = 4).