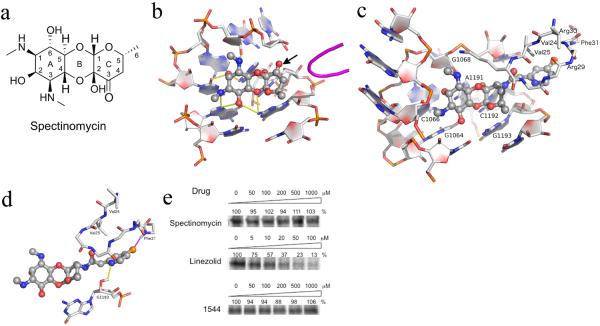
Figure 1.
(a-b) The structure and ribosomal binding site of spectinomycin. (a) Tricyclic structure of spectinomycin sketched manually with ring numbering indicated. (b) Spectinomycin bound to helix 34 with hydrogen bonding interactions indicated by yellow dashes derived from crystal structure PDB 2QOU. The nearby protein loop of RpsE is shown in magenta. From this analysis, the 3'keto functionality (indicated by arrow) on the C ring is clearly available for modification, as indicated by the arrow. (c–d) The stable conformational complex of the spectinamides in the ribosomal spectinomycin-binding site as determined by 5ns molecular dynamics simulation. (c) Compound 1544 modeled into the spectinamide helix 34 ribosomal binding site. (d) Display of the proposed binding interactions of the spectinamide side chain. E. coli numberings are shown in the figure. Corresponding numberings in M. tuberculosis for RpsE are V55(V24), V56(V25), R60(R29), R61(R30), F62(F31) and 16S are G1054(G1064), C1056(C1066), G1058(G1068), A1182(A1191), C1183(C1192), G1184(G1193). (e) In organello mitochondrial expression of S35 methionine labeled COX1 in the presence of inhibitors at indicated doses as determined by densitometry of autoradiograms. Representative results from three independent experiments are shown.