Figure 5. Linker domain structure and remodelling.
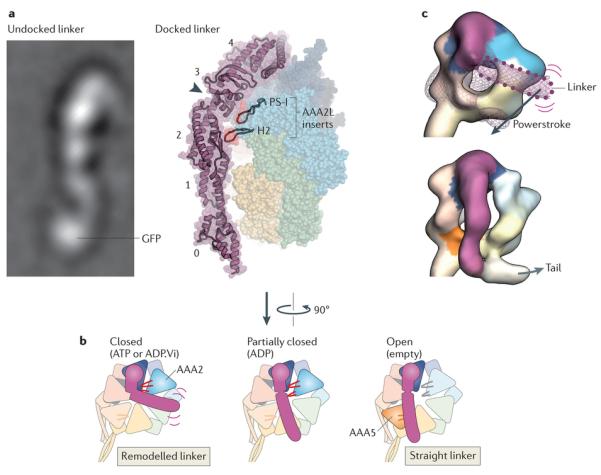
Rearrangements in the linker domain’s structure are crucial for dynein motility. a | Electron microscopy image of the linker domain from Dictyostelium discoideum dynein when it is undocked from the AAA+ ring, showing that the linker is a stable structural entity (left). A GFP tag on the amino-terminal end of the linker is labelled. Linker undocking has, thus far, been observed by electron microscopy only in purified dyneins90, 91, 92, and it is not known whether it can occur under physiological settings. View of the linker arching over the AAA+ ring in the D. discoideum dynein crystal structure in the ADP-bound state93 (right). The five subdomains in the linker are numbered (0–4). The cleft between subdomains 2 and 3, which is spanned by a single α-helix, is indicated by the arrowhead. Two β-hairpin inserts (PS-I and H2) within the large subdomain of AAA2 (AAA2L) that contact the linker near the cleft are highlighted. In addition to the cleft, note that there is a tenuous connection between linker subdomains 0 and 1 that could potentially deform, for example, under strain. b | Model of linker remodelling based on available structural data90, 91, 92, 93, 95, 118. When ATP or ADP.Vi (ADP plus vanadate) is bound in AAA1, the linker seems to be mobile (indicated by purple lines) but bent towards AAA2 in most molecules, according to cryo-EM studies of Chlamydomonas reinhardtii inner arm dynein-c and D. discoideum cytoplasmic dynein90 (left). The PS-I and H2 inserts in AAA2 (shown in red) are strong candidates to mediate this remodelling of the linker93, 95. Following inorganic phosphate (Pi) release, the linker is thought to undergo a powerstroke, in which it straightens to lie over AAA4, based on the D. discoideum dynein crystal structure93 (middle). AAA1 is partially closed around ADP, and the PS-I and H2 inserts form a limited interaction with the linker. Finally, the linker docks at AAA5, as has been observed for Saccharomyces cerevisiae cytoplasmic dynein and C. reinhardtii inner arm dynein-c in the absence of nucleotide (right). This step is proposed to fully open AAA1 and thus eject ADP95. It is unknown if the inserts within the core folds of AAA3 and AAA4 (not shown) can also interact with the linker106. c | Cryo-electron microscopy maps of C. reinhardtii inner arm dynein-c in the ADP.Vi-bound state (upper) and in the absence of nucleotide (lower)90 (Electron Microscopy Data Bank codes: 2156 and 2155, respectively). The maps are coloured as in part b, with AAA+ modules interacting with the linker shown in saturated colours and other modules in pale colours. In the ADP.Vi map, density for the distal linker is missing, owing to variability in its position. The position of the distal linker suggested by a variance map (purple wiremesh) and GFP-based tagging is shown with a dotted outline90. Straightening of the linker is thought to represent the powerstroke of dynein (Fig. 3). Electron microscopy image in part a courtesy of B. Malkova, Biomolecular Research Laboratory, Paul Scherrer Institute, Switzerland.
