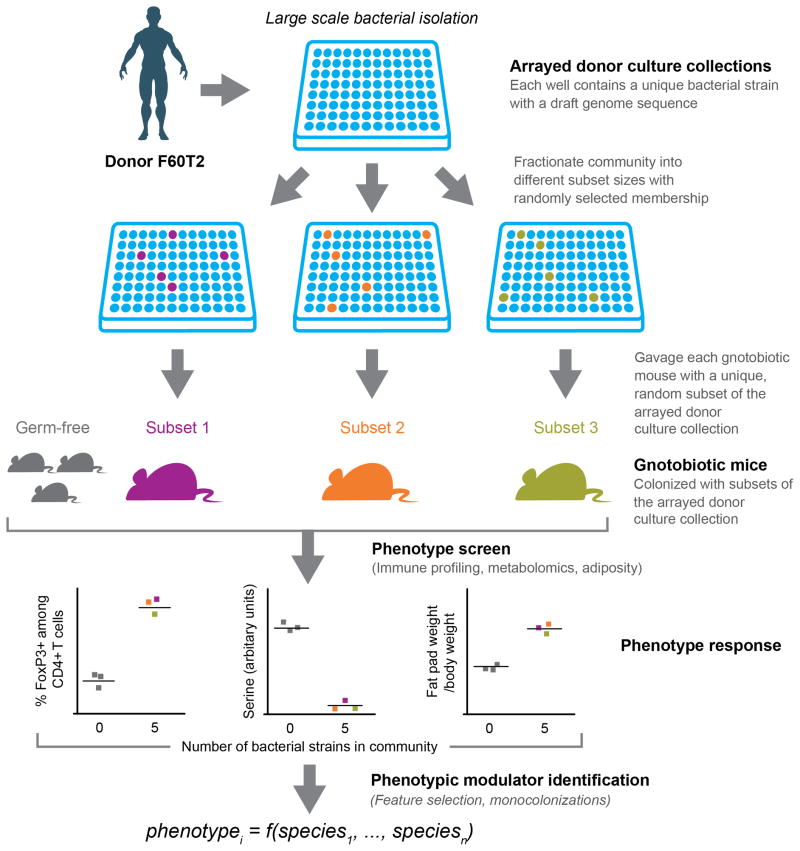
Fig. 1. Overview of combinatorial ‘out-of-the-isolator’ gnotobiotics screen for identifying microbe-phenotype relationships.
Bacterial culture collections are generated from an intact uncultured (human) gut microbiota sample whose transplantation into gnotobiotic mice has already been shown to alter the animal’s physiologic, metabolic, immunologic or other properties. Each strain in the culture collection is present in a separate well of a multi-well plate. The arrayed collection of sequenced bacterial strains is then fractioned into random subsets of various sizes [shown are subsets (consortia) of five strains]. Each subset is gavaged into an individual germ-free animal maintained in a separate filter-top gnotobiotic cage to observe the effect of the subset on phenotypes of interest. By repeating this process across many subsets, the effect of each strain in the arrayed culture collection is assayed in the context of a diverse background of community memberships and sizes. Feature selection algorithms (for non-saturated phenotypes) and follow-up mono-colonization experiments (for phenotypes saturating at small community sizes) are used to identify the strains whose presence or absence best explains the observed phenotypic variation.