Abstract
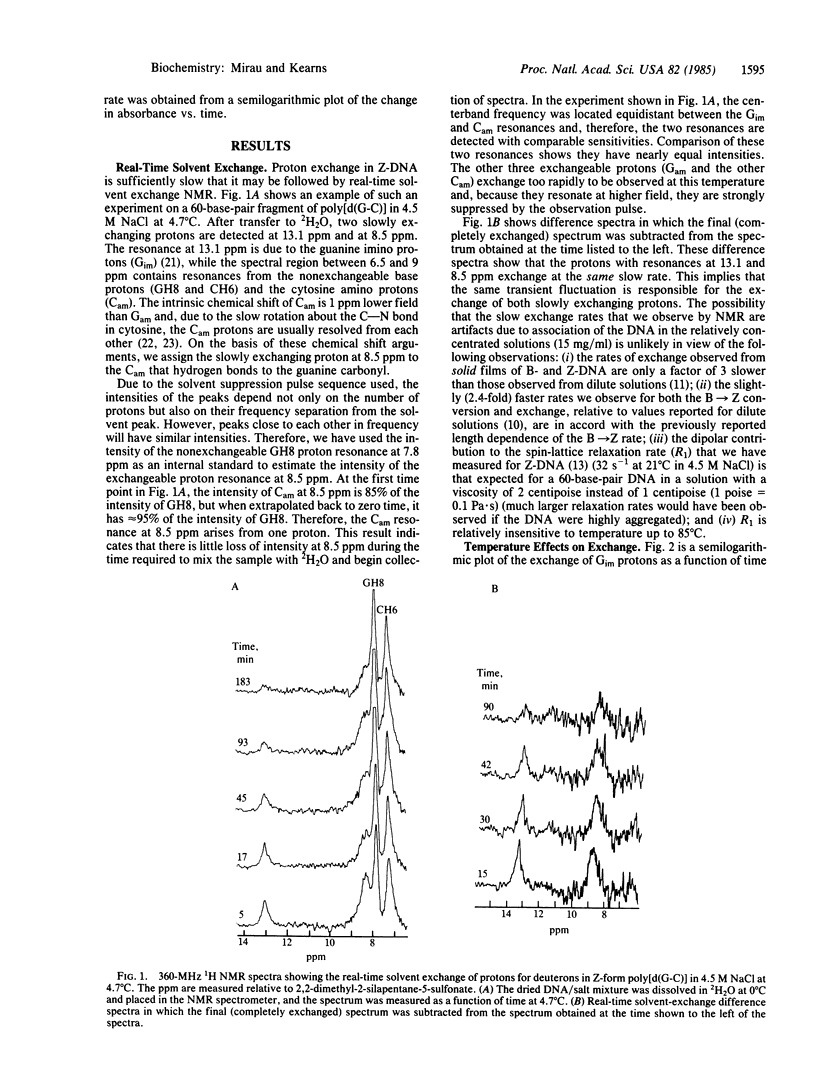
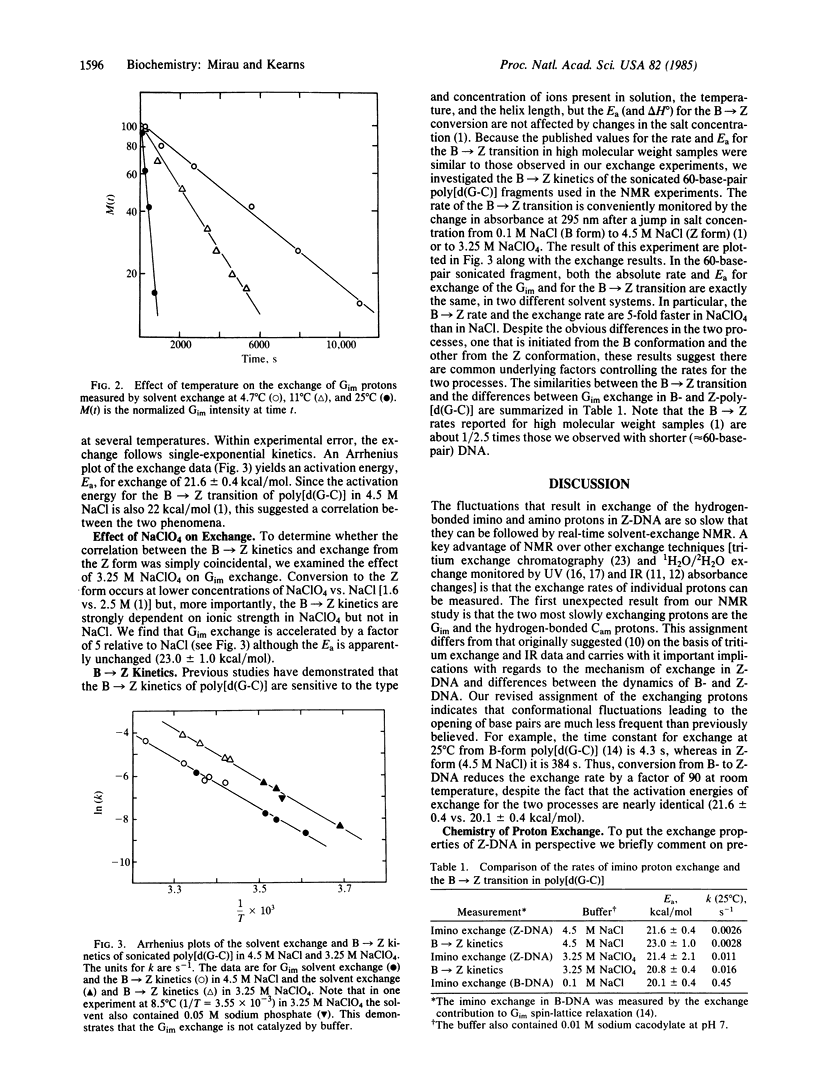
Real-time solvent-exchange NMR has been used to study the proton exchange in Z-form poly[d(G-C)]. In 4.5 M NaCl the most slowly exchanging protons (about two orders of magnitude slower than in B-DNA) are identified as the guanine imino proton and the cytosine amino proton hydrogen bonded to the guanine carbonyl. Both protons exchange at the same rate and the exchange follows single-exponential kinetics and cannot be catalyzed, implying that the exchanges of the two protons both occur from the same transient solvent-exposed state. The exchange depends strongly on temperature and the activation energy for exchange (≈22 kcal/mol) is the same as the activation energy for the B → Z transition. The rate of proton exchange is identical with the B → Z transition rate, both measured in 4.5 M NaCl. The correlation between the B → Z kinetics and the proton exchange also extends to 3.25 M NaClO4 solutions, in which both rates are 5 times faster. This unexpected parallelism between the B → Z transition kinetics and the Z exchange kinetics indicates that the rate-limiting steps in the two processes are similar.
Keywords: NMR, DNA “breathing”, B → Z kinetics, DNA dynamics
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Assa-Munt N., Granot J., Behling R. W., Kearns D. R. 1H NMR relaxation studies of the hydrogen-bonded imino protons of poly(dA-dT). Biochemistry. 1984 Feb 28;23(5):944–955. doi: 10.1021/bi00300a023. [DOI] [PubMed] [Google Scholar]
- Drew H., Takano T., Tanaka S., Itakura K., Dickerson R. E. High-salt d(CpGpCpG), a left-handed Z' DNA double helix. Nature. 1980 Aug 7;286(5773):567–573. doi: 10.1038/286567a0. [DOI] [PubMed] [Google Scholar]
- Glickson J. D., Phillips W. D., Rupley J. A. Proton magnetic resonance study of the indole NH resonances of lysozyme. Assignment, deuterium exchange kinetics, and inhibitor binding. J Am Chem Soc. 1971 Aug 11;93(16):4031–4038. doi: 10.1021/ja00745a035. [DOI] [PubMed] [Google Scholar]
- Hartmann B., Pilet J., Ptak M., Ramstein J., Malfoy B., Leng M. The B reversible Z transition of poly(dI-br5dC).poly(dI-br5dC). A quantitative description of the Z form dynamic structure. Nucleic Acids Res. 1982 May 25;10(10):3261–3277. doi: 10.1093/nar/10.10.3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. D., Figueroa N., Redfield A. G. Real-time solvent exchange studies of the imino and amino protons of yeast phenylalanine transfer RNA by Fourier transform NMR. Proc Natl Acad Sci U S A. 1979 Jul;76(7):3130–3134. doi: 10.1073/pnas.76.7.3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns D. R. High-resolution nuclear magnetic resonance studies of double helical polynucleotides. Annu Rev Biophys Bioeng. 1977;6:477–523. doi: 10.1146/annurev.bb.06.060177.002401. [DOI] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Nuclear Overhauser experiments at 500 MHz on the downfield proton spectrum of a ribonuclease-resistant fragment of 5S ribonucleic acid. Biochemistry. 1983 May 24;22(11):2615–2622. doi: 10.1021/bi00280a004. [DOI] [PubMed] [Google Scholar]
- Kłysik J., Stirdivant S. M., Larson J. E., Hart P. A., Wells R. D. Left-handed DNA in restriction fragments and a recombinant plasmid. Nature. 1981 Apr 23;290(5808):672–677. doi: 10.1038/290672a0. [DOI] [PubMed] [Google Scholar]
- Lafer E. M., Möller A., Nordheim A., Stollar B. D., Rich A. Antibodies specific for left-handed Z-DNA. Proc Natl Acad Sci U S A. 1981 Jun;78(6):3546–3550. doi: 10.1073/pnas.78.6.3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal C., Kallenbach N. R., Englander S. W. Base-pair opening and closing reactions in the double helix. A stopped-flow hydrogen exchange study in poly(rA).poly(rU). J Mol Biol. 1979 Dec 5;135(2):391–411. doi: 10.1016/0022-2836(79)90443-1. [DOI] [PubMed] [Google Scholar]
- McConnell B. The amino 1H resonances of oligonucleotide helices: d(CGCG). J Biomol Struct Dyn. 1984 Jun;1(6):1407–1421. doi: 10.1080/07391102.1984.10507528. [DOI] [PubMed] [Google Scholar]
- Mirau P. A., Kearns D. R. Effect of environment, conformation, sequence and base substituents on the imino proton exchange rates in guanine and inosine-containing DNA, RNA, and DNA-RNA duplexes. J Mol Biol. 1984 Aug 5;177(2):207–227. doi: 10.1016/0022-2836(84)90453-4. [DOI] [PubMed] [Google Scholar]
- Nakanishi M., Tsuboi M. Two channels of hydrogen exchange in a double-helical nucleic acid. J Mol Biol. 1978 Sep 5;124(1):61–71. doi: 10.1016/0022-2836(78)90147-x. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Lafer E. M., Peck L. J., Wang J. C., Stollar B. D., Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982 Dec;31(2 Pt 1):309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- Nordheim A., Pardue M. L., Lafer E. M., Möller A., Stollar B. D., Rich A. Antibodies to left-handed Z-DNA bind to interband regions of Drosophila polytene chromosomes. Nature. 1981 Dec 3;294(5840):417–422. doi: 10.1038/294417a0. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Nordheim A., Rich A., Wang J. C. Flipping of cloned d(pCpG)n.d(pCpG)n DNA sequences from right- to left-handed helical structure by salt, Co(III), or negative supercoiling. Proc Natl Acad Sci U S A. 1982 Aug;79(15):4560–4564. doi: 10.1073/pnas.79.15.4560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilet J., Leng M. Comparison of poly(dG-dC).poly(dG-dC) conformations in oriented films and in solution. Proc Natl Acad Sci U S A. 1982 Jan;79(1):26–30. doi: 10.1073/pnas.79.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohl F. M., Jovin T. M. Salt-induced co-operative conformational change of a synthetic DNA: equilibrium and kinetic studies with poly (dG-dC). J Mol Biol. 1972 Jun 28;67(3):375–396. doi: 10.1016/0022-2836(72)90457-3. [DOI] [PubMed] [Google Scholar]
- Ramstein J., Leng M. Salt-dependent dynamic structure of poly(dG-dC) x poly(dG-dC). Nature. 1980 Nov 27;288(5789):413–414. doi: 10.1038/288413a0. [DOI] [PubMed] [Google Scholar]
- Robert-Nicoud M., Arndt-Jovin D. J., Zarling D. A., Jovin T. M. Immunological detection of left-handed Z DNA in isolated polytene chromosomes. Effects of ionic strength, pH, temperature and topological stress. EMBO J. 1984 Apr;3(4):721–731. doi: 10.1002/j.1460-2075.1984.tb01875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoup R. R., Miles H. T., Becker E. D. NMR evidence of specific base-pairing between purines and pyrimidines. Biochem Biophys Res Commun. 1966 Apr 19;23(2):194–201. doi: 10.1016/0006-291x(66)90527-4. [DOI] [PubMed] [Google Scholar]
- Teitelbaum H., Englander S. W. Open states in native polynucleotides. I. Hydrogen-exchange study of adenine-containing double helices. J Mol Biol. 1975 Feb 15;92(1):55–78. doi: 10.1016/0022-2836(75)90091-1. [DOI] [PubMed] [Google Scholar]
- Teitelbaum H., Englander S. W. Open states in native polynucleotides. II. Hydrogen-exchange study of cytosine-containing double helices. J Mol Biol. 1975 Feb 15;92(1):79–92. doi: 10.1016/0022-2836(75)90092-3. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]


