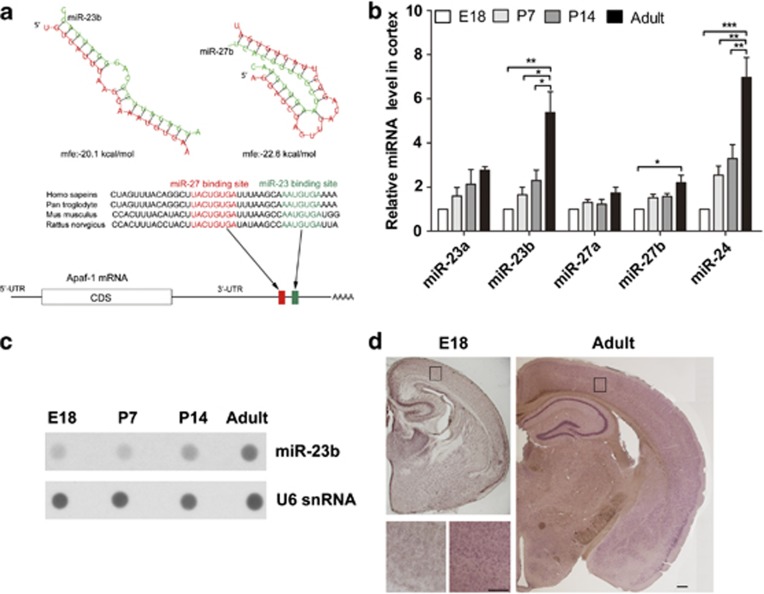
Figure 2.
The expression of miR-23-27 clusters increases during brain development. (a) Schematic of the Apaf-1 3′UTR indicating the locations of the miR-23 and miR-27 target sites that are conserved in vertebrates. The Apaf-1 3′UTR contains evolutionarily well-conserved sequences matched for the miR-23 and miR-27 families that were predicted by computer-aided algorithms. The free energies (mfes) of microRNA bindings were calculated by RNAHybrid software (BiBiServ, Bielefeld, Germany). (b) Quantitative RT-PCR detection of miR-23a, miR-23b, miR-27a, miR-27b, and miR-24 in cerebral cortex samples at different developmental stages (n=5, one-way ANOVA with Newman–Keuls multiple comparison test, *P<0.05, **P<0.01, and ***P<0.001). (c) Dot blot analyses of miR-23b and U6 snRNA in total RNA of the cerebral cortex. Each lane was loaded with equal amounts of total RNA extracted from the mixed samples of five individuals. (d) Expression of miR-23b in cortices of E18 and adult brain tissues detected by in situ hybridization. Scale bar represents 250 μm; scale bar (insert) represents 200 μm