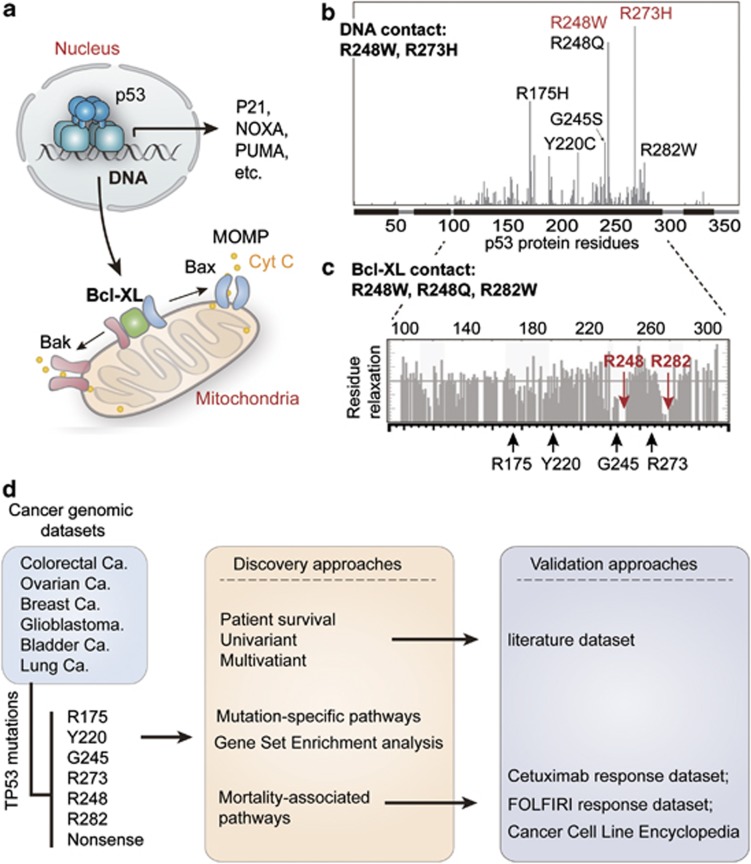
Figure 1.
Classification of p53 mutations and study design. (a) The function of p53 in nucleus and mitochondria. In response to DNA damage, p53 functions as a transcription factor, triggering cell cycle arrest and apoptosis by transactivation of p21, Noxa, Puma, so on. In addition, the function of p53 in mitochondria is mediated by its interaction with Bcl-XL, which releases Bak/Bax and causes mitochondrial outer membrane permeabilization (MOMP)-induced leakage of cytochrome c (cyt c) and apoptosis. (b) Frequently-occuring (hotspot) p53 mutations and classification of p53 mutations according to its effects on DNA contact and protein structural stability. The x-axis indicates amino acid sequence of p53 protein, and the y-axis displays frequency of mutation in the TCGA data sets. DNA contact mutations are labeled in red, whereas structural mutations are marked in black. (c) The position of hotspot mutations in relation to the regions of p53 that contact Bcl-XL. The gray bar plot indicates relaxation of p53 residues while in contact with Bcl-XL (higher relaxation indicates noncontact residues) as determined by nuclear magnetic resonance previously.11 The mutated residues sitting in the contact regions (R248 and R282) are highlighted in red. (d) Schematic diagram representing the study processes. Cancer cases collected from the indicated data sets of The Cancer Genome Atlas (TCGA) were grouped according to p53 mutations. The patient survival was compared using univariant and multivariant models, and then validated using an independent data set extracted from literatures. The enriched (signature) pathways for different mutations were determined using microarray data of TCGA colorectal cancer data set. Finally, the effects of p53 mutation and expression of CYP3A4 in response to FOLFIRI and cetuximab were analyzed using the indicated data sets