Figure 1.
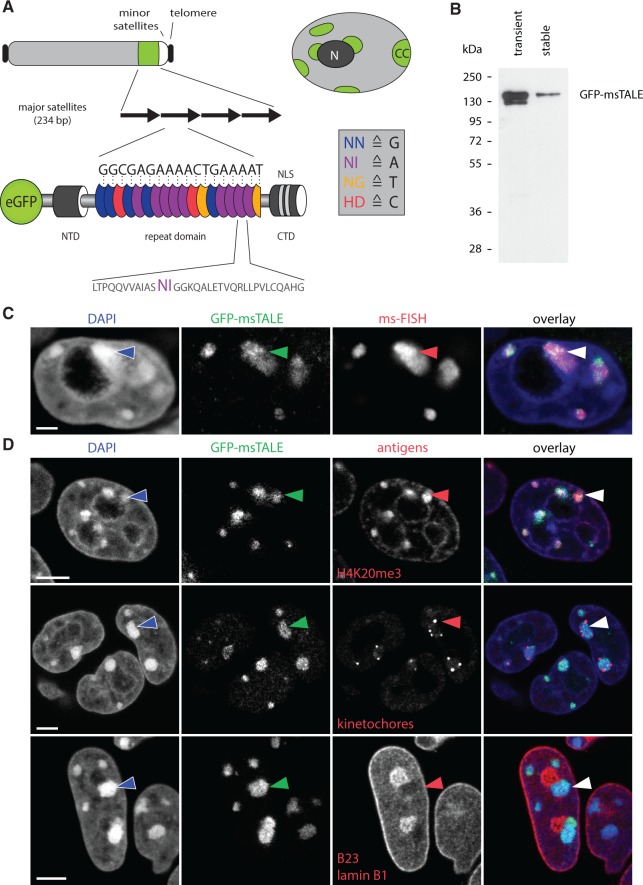
Localization of the GFP-msTALE to major satellite repeats in mouse pericentromeric heterochromatin. (A, top) Schematic representation of an acrocentric mouse chromosome with telomeres (black), major satellites (green), minor satellites (white) and the long arm of the chromosome (light gray). Overview of a nucleus showing multiple heterochromatin centers (CC, green), where the major satellite DNA is clustered. CCs localize next to the nuclear periphery and the nucleoli (dark gray, N) and are surrounded by less condensed chromatin (light gray). (A, bottom) Schematic representation of the GFP-msTALE aligned to its binding site within the major satellite repeats (black arrows). The dTALE is composed of an N-terminal domain (NTD), a C-terminal domain (CTD) bearing nuclear localization signals (NLS) and a central repeat domain. DNA target recognition is mediated by the RVDs within each TALE repeat (blue, purple, yellow and red ellipses for RVDs binding to the bases G, A, T and C, respectively, single letter code for amino acids and nucleotide bases). A representative repeat sequence with RVDs (purple) is shown below as close-up (single letter code for amino acids). The complete sequence is shown in Supplementary Figure S1. Note that the msTALE is lacking the C-terminal activation domain. For visualization and immunoprecipitation, the dTALE is N-terminally fused to GFP. (B) GFP-Trap pull-down from HEK293T cells transiently transfected with the GFP-msTALE construct (transient) and a J1 ESC clone stably expressing the GFP-msTALE (stable). Immunodetection by an anti-GFP antibody. (C) 3D-immuno-FISH on stable GFP-msTALE ESCs with probe directed against major satellite repeats (ms-FISH). Because the GFP signal is strongly reduced by 3D-FISH procedure, an anti-GFP antibody was used to visualize GFP-msTALE localization. Note strict co-localization of the GFP signal (green) and the FISH probe (red). Nuclei were counterstained with DAPI (blue). Arrowhead points at one of the CCs. (D) Immunostaining of ESCs stably expressing the GFP-msTALE (green). Upper panel, antibodies against heterochromatin (anti-H4K20me3, red) mark GFP-positive CCs. Middle panel, human antiserum binding to kinetochores reveals kinetochore clusters (red) at the surface of CCs. Lower panel, CCs marked with GFP-msTALE show a characteristic intranuclear localization abutting nuclear periphery or adjacent to the nucleoli (both shown in red). Nuclei were counterstained with DAPI (blue). Arrowheads mark one of the CCs in each exemplified nucleus. All images are single optical confocal sections. Scale bars: C, 5 µm; D, 2 µm.