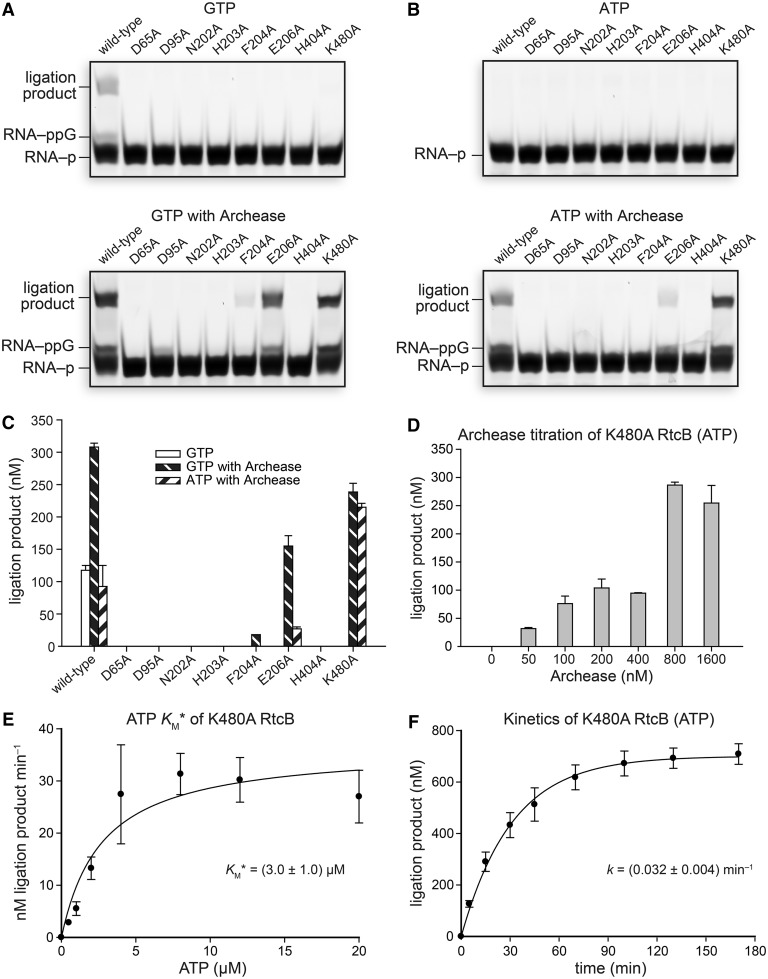
Figure 7.
Effect of Archease on the activity of active-site variants of RtcB in RNA ligation assays with GTP or ATP as a cofactor. (A) Reactions with GTP (0.10 mM) as a cofactor. (B) Reactions with ATP (0.10 mM) as a cofactor. Reaction mixtures included 100 nM Archease where indicated, and were incubated at 70°C for 30 min. (C) Graph of the ligation product obtained for each RtcB variant. Values are the mean ± SE for two separate experiments. (D) ATP-dependent K480A RtcB-catalyzed RNA ligation reactions titrated with increasing concentrations of Archease, as specified. ATP was included at 0.10 mM, and reaction mixtures were incubated at 70°C for 15 min. Values are the mean ± SE for three separate experiments. (E) Michaelis–Menten plot of reaction rate versus ATP cofactor concentration for K480A RtcB-catalyzed RNA ligation reactions under single-turnover conditions. Values are the mean ± SE for three separate experiments. (F) Single-turnover kinetics of ATP-dependent RNA ligation catalyzed by K480A RtcB with the inclusion of Archease (800 nM). Values are the mean ± SE for two separate experiments. Ligation reaction mixtures contained 50 mM Bis–Tris buffer (pH 7.0), NaCl (300 mM), MnCl2 (0.25 mM), NTP as indicated, P. horikoshii RtcB (5 μM), 5′ RNA fragment (1.0 μM) and 3′ RNA fragment (1.0 μM).