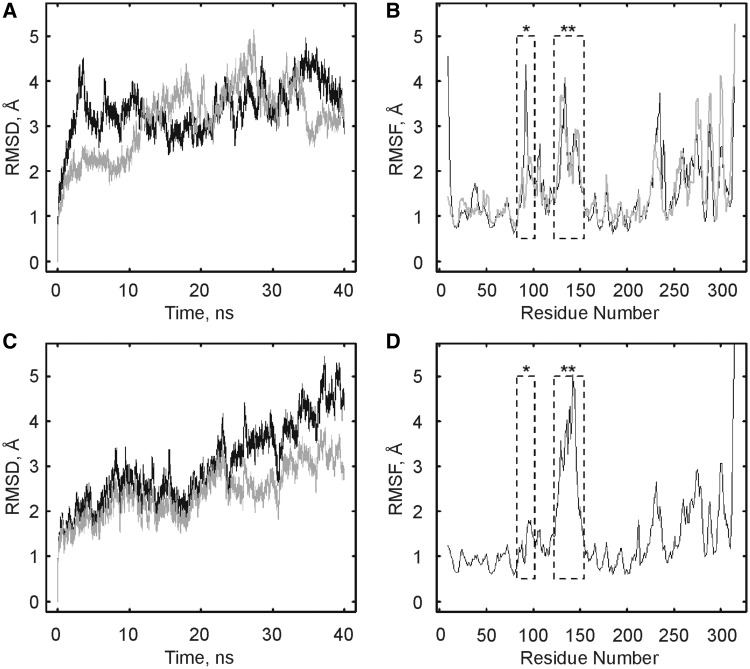
Figure 6.
Global fluctuations of TruB during MD simulations. TruB wild type (in the absence of RNA, panels (A and B) was simulated at 310 K in the apo and bound conformation based on available crystal structures with and without RNA, respectively (for details see ‘Materials and Methods’ section). TruB D90N (C and D) was simulated in the bound conformation only. (A) RMSD of the backbone atoms of all residues for TruB wild type simulations (apo conformation, gray; bound conformation, black). (B) RMSF values for simulation of TruB wild type from 5–40 ns, with insert 1 (asterisk) and thumb-loop (double asterisk) residues indicated by dashed boxes (apo conformation, gray; bound conformation, black). (C) TruB D90N simulation. RMSD of the backbone atoms of all residues (black) and for backbone atoms of all residues excluding thumb-loop residues 124–152 (gray). (D) RMSF values for TruB D90N residues from 5 to 40 ns of simulation; insert 1 (asterisk) and thumb loop (double asterisk).