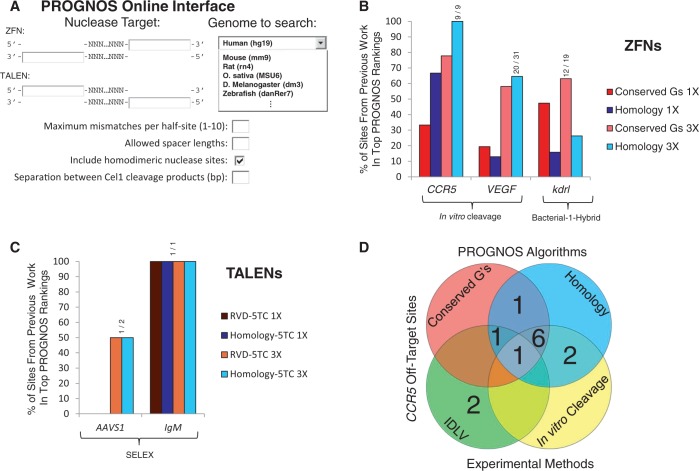
Figure 1.
PROGNOS search interface and comparison to previous prediction methods. (A) The PROGNOS online interface allows users to enter the target site of their nuclease pair and specify search parameters and primer design considerations. (B) A comparison of PROGNOS predictions to previously reported methods identifying off-target sites for different ZFNs (6,9). The Homology and Conserved G’s algorithms were used to determine what percentage of the sites with previously identified off-target activity fell within the top fractions of PROGNOS rankings. The ‘1X’ top fraction corresponds to searching the same number of top PROGNOS sites as were investigated in the original paper and ‘3X’ corresponds to searching three times as many PROGNOS sites as were investigated in the original manuscript. (C) A comparison of the PROGNOS search algorithms to previously reported methods identifying off-target sites for TALENs (5,8). The top PROGNOS rankings using the Homology-5TC and RVD-5TC algorithms were searched to determine what percentage of off-target sites found to have activity fell within the top fractions of PROGNOS rankings. (D) Venn diagram displaying the 13 known off-target sites identified for the heterodimeric CCR5 ZFNs during development and testing of the original PROGNOS algorithms (9,10). The sites ranked at the top of the PROGNOS Homology and Conserved G’s in silico algorithms [allowing 3X the number of sites searched by Pattanayak et al. (9)] are compared to the 12 sites identified previously and one site uncovered in this study.