Figure 5.
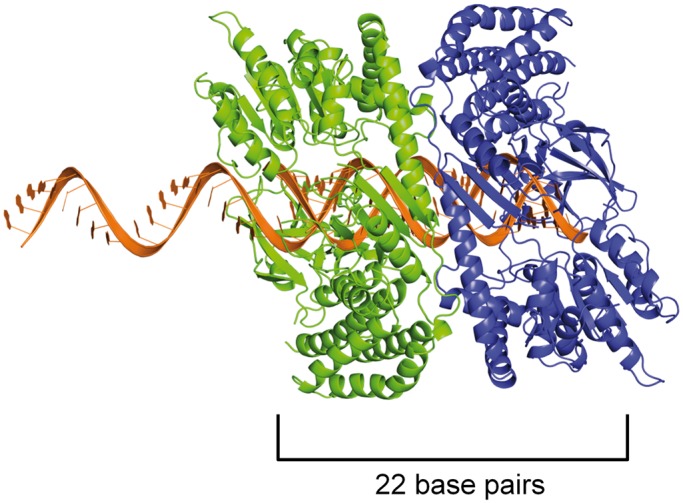
Model of a helicase domain dimer on an mRNA/22G-RNA duplex. As a proxy for the closely related DRH-3 protein, we used the crystal structure of the RIG-I helicase domain (11) and modeled it bound to a 22-bp duplex. Because 22G-RNA is complementary to a longer mRNA target, we imagine that the formation of the DRH-3 dimer occurs immediately after synthesis, when the mRNA and 22G-RNA are in duplex form. The top strand is thus extended beyond the duplex in the 5′ direction, as 22G-RNA is complementary to upstream mRNA target sequences that have yet to be synthesized. We show one protein monomer (in blue) interacting with the 5′ G of the 22G-RNA strand via the C-terminal domain, as is presented in the RIG-I structure, and because we show here that ΔN-DRH3 can recognize 5′-triphosphates. When a second protein molecule (in green) is bound adjacent to the first, the footprint is roughly 22 bp in size, thus creating a means for measurement by the dimer. The protein monomers are oriented such that the Hel2 domains are in close proximity, which is supported by the observation that the DRH-3 dimer is necessary for efficient ATP hydrolysis.
