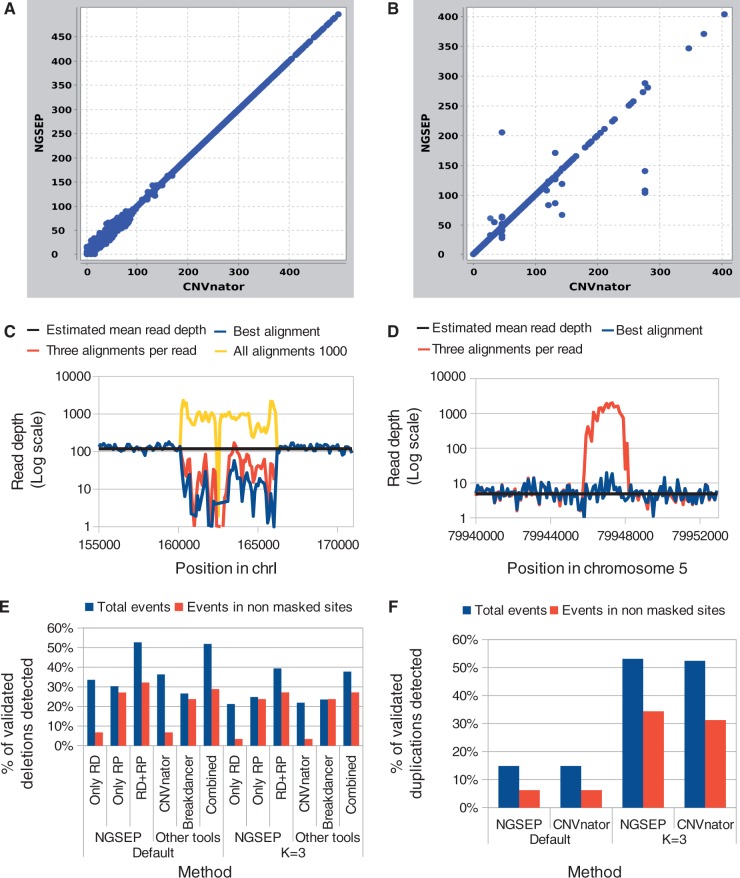
Figure 4.
Quality assessment of the implementation of the CNVNator algorithm in NGSEP. Given the same GC-corrected intensities, the same genome size and the same RD distribution parameters, both implementations produce nearly the same (A) partition and (B) RD levels. Examples of repetitive regions in (C) the yeast parent ER7A and (D) the low-coverage human sample show how RD varies depending on the number of alignments counted for each read (blue: only the best alignment of each read counted; red: up to three alignments counted; yellow: all alignments found with Bowtie 2 with the -a option counted). Sensitivity of NGSEP, CNVnator and BreakDancer to identify (E) deletions and (F) duplications validated by Mills and collaborators (22) using reads from the low-coverage data set for NA12878. Default and K = 3 modes of Bowtie 2 are compared.