Figure 6.
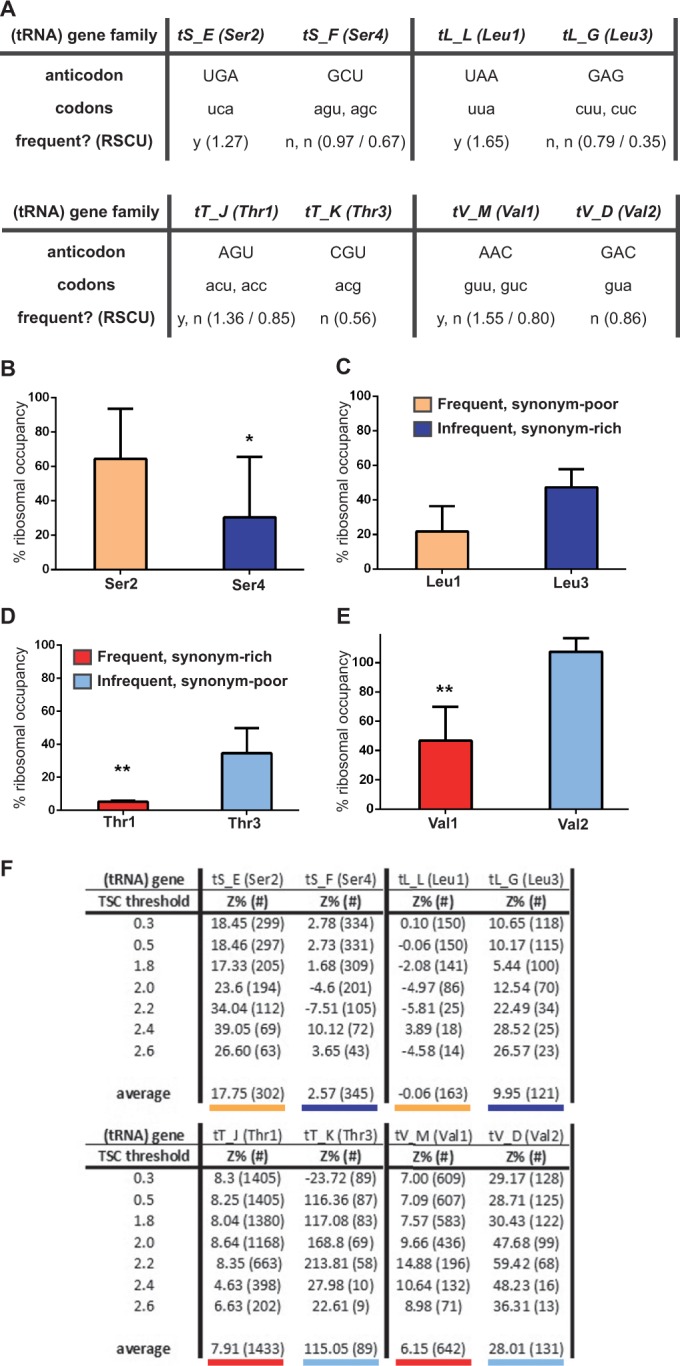
The tRNAs recognizing codons less autocorrelated in Scp160p target mRNAs are more depleted from ribosomes following Scp160 depletion. This effect in independent both of the number of synonymous codons, or their frequency in the genome. (A) The tRNA gene family names, common tRNA names, anticodon used, codons recognized and RSCU for the candidate pairs contrasting within the four amino acids experimentally assessed. (B–E) Residual ribocc of tRNA pairs for amino acids on Scp160p depletion, displayed as percentage of wild-type levels. The standard deviation between at least two biological replicates with two technical replicates each is indicated, while *, P <0.1 and **, P <0.01 for significance of independent average values between knockdown and wild-type (above columns). (B) Serine, comparing Ser2 and Ser4: tRNAs recognizing only one frequent (orange) versus several infrequent (dark blue) codons. (C) Leucine, comparing Leu1 and Leu3: tRNAs recognizing only one frequent versus several infrequent codons. (D) Threonine, comparing Thr1 and Thr3: tRNAs recognizing several frequent (red) versus only one infrequent (light blue) codon. (E) Valine, comparing Val1 and Val2: tRNAs recognizing several frequent versus only one infrequent codon. (F) Z-score percentage (equals to codon autocorrelation at the tRNA level) in mRNA target subgroups ordered by TSC (equivalent to degree of mRNA distribution shift in polysome gradients) threshold. Z-scores were calculated individually for each indicated set up to the indicated TSC threshold. All tRNAs analyzed are shown. For enhanced clarity, underlinings of average Z-score values correspond to color-coding of the respective tRNA species analyzed experimentally in the above figures, with red versus blue shades for popular or unpopular codons, and bold versus faint shades for synonym-rich or synonym-poor tRNAs.
