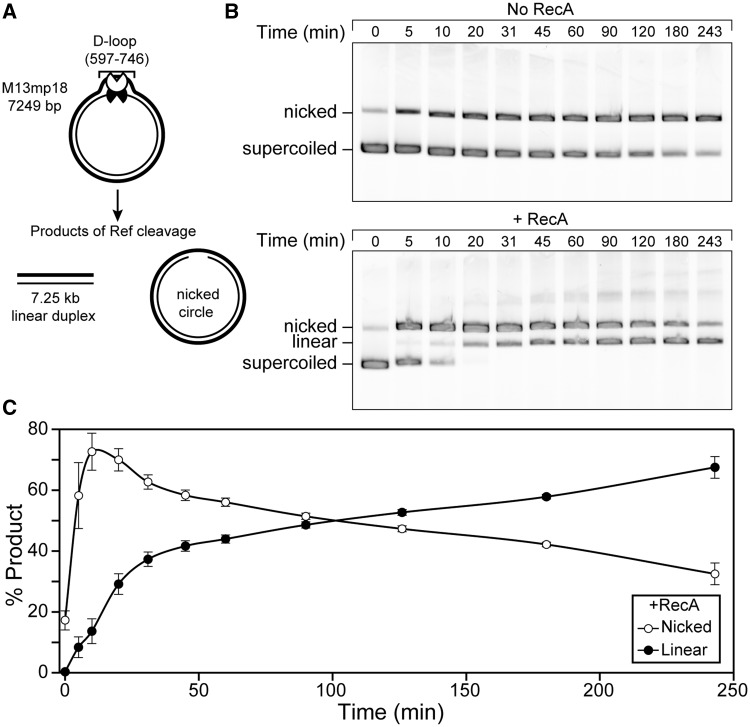
Figure 1.
Ref creates targeted DSBs in cdsDNA through a nicked intermediate. (A) The reaction scheme is shown. RecA E38K (1.33 µM) was incubated with rlb1 oligonucleotide (4 µMnt) in buffer A* (8.5, 15) with an ATP regeneration system for 10 min to form a RecA filament. ATP (3 mM) was added and incubated for 20 min, followed by the addition of M13mp18 cdsDNA (8 µMnt). This was incubated for 20 min and a D-loop structure formed at positions 597–746 in the dsDNA (shown). Ref (100 nM) was added, and DNA was cleaved within the D-loop. One nick by Ref results in nicked cdsDNA, whereas two cuts result in linear dsDNA. (B) Agarose gel representing time points from the targeted nuclease assay, without and with (top and bottom, respectively) RecA E38K present. Supercoiled DNA is the starting material (bottom band), which is converted first to nicked DNA (top band) and subsequently to linear DNA (middle band). A small amount of RecA-independent nuclease activity is seen (top). (C). Graphical representation of (B; bottom) averaged over three independent experiments. Nicked product (open circles) appears quickly, and then disappears as it is converted to linear product (closed circles).