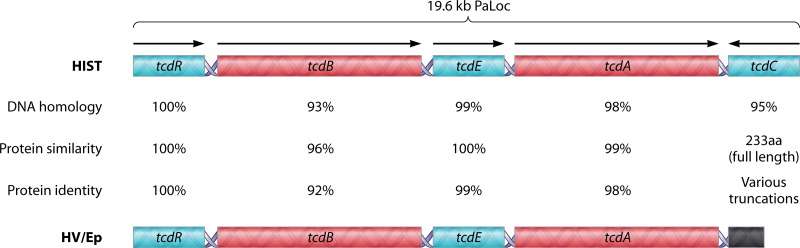
Fig 2.
Comparison of C. difficile pathogenicity loci between historical and hypervirulent strains. The schematics depict the 19.6-kb PaLoc of historical C. difficile (HIST) (top) and hypervirulent C. difficile (HV/Ep) (bottom). DNA homology, protein similarity, and protein identity are based upon reference strains CD630 (GenBank accession no. NC_009089.1) and BI1 (GenBank accession no. NC_017179.1). TcdC protein similarity and identity calculations are not trivial, as truncations of tcdC vary among hypervirulent/epidemic (HV/Ep) isolates, resulting in final products ranging from 61 to 226 amino acids (aa).