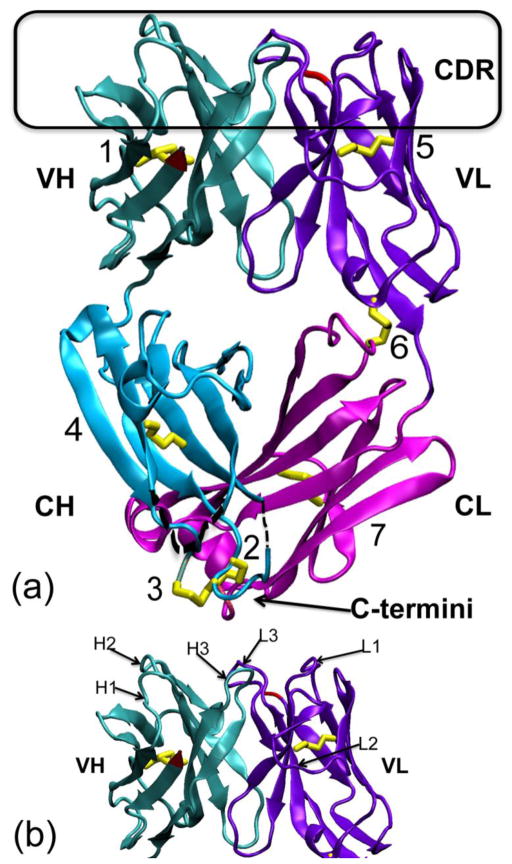
Figure 1. Crystal structure of the M204 antigen-binding fragment (Fab).
(a) The 204Fab structure (PDB ID: 3NL4) in ribbon representation. The four domains of the Fab, the variable (V) and constant (C) domains of the heavy (H) and light (L) chains, are noted as VH (teal), CH (aqua), VL (violet), and CL (magenta), respectively. The complementarity determining region (CDR) loops are boxed in black at the top of the figure. The N-termini of both chains are depicted in red in the VH and VL domain. The C-termini of the both chains are very close to each other and are shown at the bottom of the figure, noted with an arrow. The seven disulfide bonds (1. H-Cys21:H-Cys92, 2. H-Cys129:L-Cys213, 3. H-Cys130:H-Cys215, 4. H-Cys142:H-Cys195, 5. L-Cys23:L-Cys88, 6. L-Cys80:L-Cys172, and 7. L-Cys136:L-Cys195) are highlighted in yellow. Of these, the disulfide bond, rabbit unique disulfide bonds are boxed in orange on the left side and at the bottom of the figure. These disulfide bonds are shown and described in more details in Figure 2. Both alternative conformations of the disulfide bond between H-Cys21 and H-Cys92 are shown in the VH domain. Two short regions not modeled in the CH domain, from H-135 to H-136 (2 residues) and from H-189 to H-191 (3 residues), are denoted with black dotted lines. (b) The figure shows each individual CDR loop with respect to the variable domains of the H and L chains indicated as VH and VL, respectively. CDR-H1, -H2, -H3 and CDR-L1, -L2, -L3 are labeled H1, H2, H3 and L1, L2, L3, respectively.