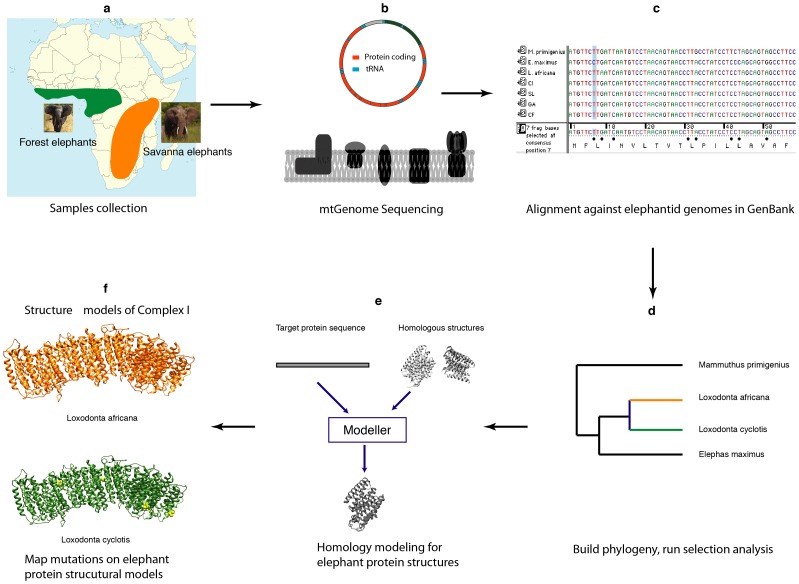
Figure 1. Flowchart outlining the methodological steps taken in our integrative approach to identify and analyze the structural biology of sites in the mitochondrial genome under positive selection in the African elephant.
(a) Sample collection; green shows the range of the forest elephant (L. cyclotis) and orange shows the range of the savanna elephant (L. africana). (b) Sequencing the mtGenome; the protein coding genes encode for the subunits of the complexes involved in OXPHOS as shown in cartoon form. (c) Sequence alignment; complete mtGenome sequences for members of the Elephantidae were downloaded from GenBank, and to which we aligned our novel forest elephant sequences. (d) Phylogenetic and selection analyses; we inferred a phylogeny from our complete, aligned mtGenome sequence data and used the output to run analyses identifying sites that might be under positive selection. (e) Homology protein modeling; after identifying which genes (and complexes) might have sites under position selection, we searched the Protein Data Bank for homologous crystal structures, then input our elephant sequences and used Modeller to predict the elephant protein structures. (f) Mutation mapping; lastly, we mapped the residues that might be under positive selection onto our predicted elephant protein structures and assessed what impacts those substitutions found between L. cyclotis (green) and L. africana (orange) might have on the function of the protein in order to relate that to biological differences.